
Matteo RAUZI
Morphogenesis and mechanics of epithelial tissues
Main interests
- Uncovering the fundamental cell working principles and the emerging supracellular mechanisms driving epithelial morphogenesis
- Unraveling the biomechanical force fields directing flow and change in shape of epithelial tissues
- Understanding how patterns of gene expression result in tissue shape transformations during embryo development
Scientific Questions
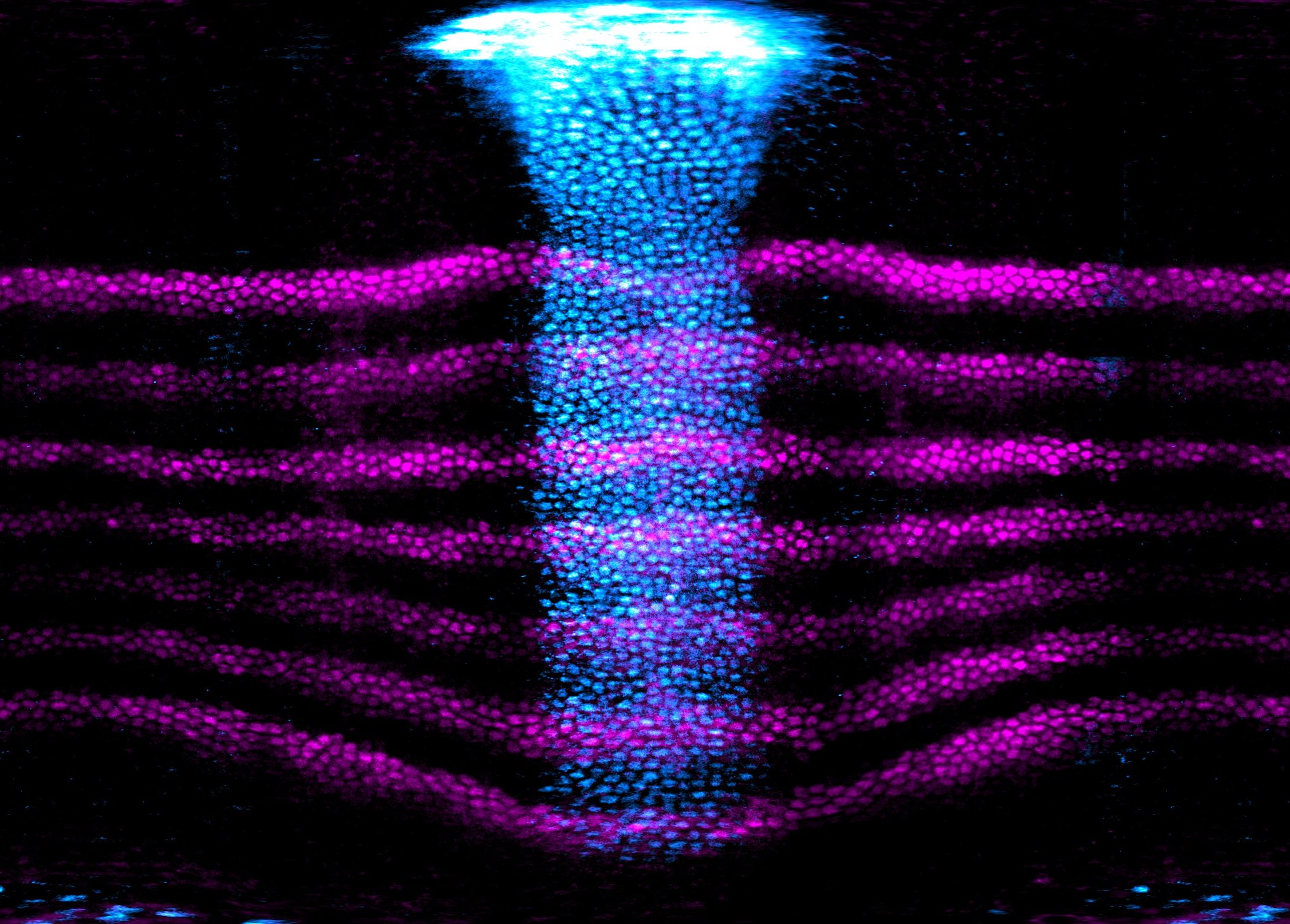
Morphogenesis builds living shapes. The change in shape of a cell is controlled by molecular signals and it is driven by mechanical forces powered by the cell cytoskeleton. Therefore, cell shape changes rely on the transfer and conversion of biochemical and mechanical energy. While key molecular players, signaling pathways and the cellular mechanics have been tested and deciphered, it is still unclear how these work at larger spatial scales. In our lab we are focused on understanding how cellular and sub-cellular properties are integrated at the embryo scale to give rise to conserved emerging mechanisms necessary to drive coordinated tissue flows and remodeling during development. To that end we apply a spectrum of cutting edge imaging, molecular and biophysical techniques and tools to tackle morphogenesis from the molecule to the embryo.
Figure: expression pattern of the dorsal-ventral patterning gene snail (blue) and of the anterior-posterior patterning gene eve (magenta) shown over a Drosophila embryo cylindrical projection. Ventral in the center, dorsal on the right and left, anterior top and posterior bottom.
Our Strategy
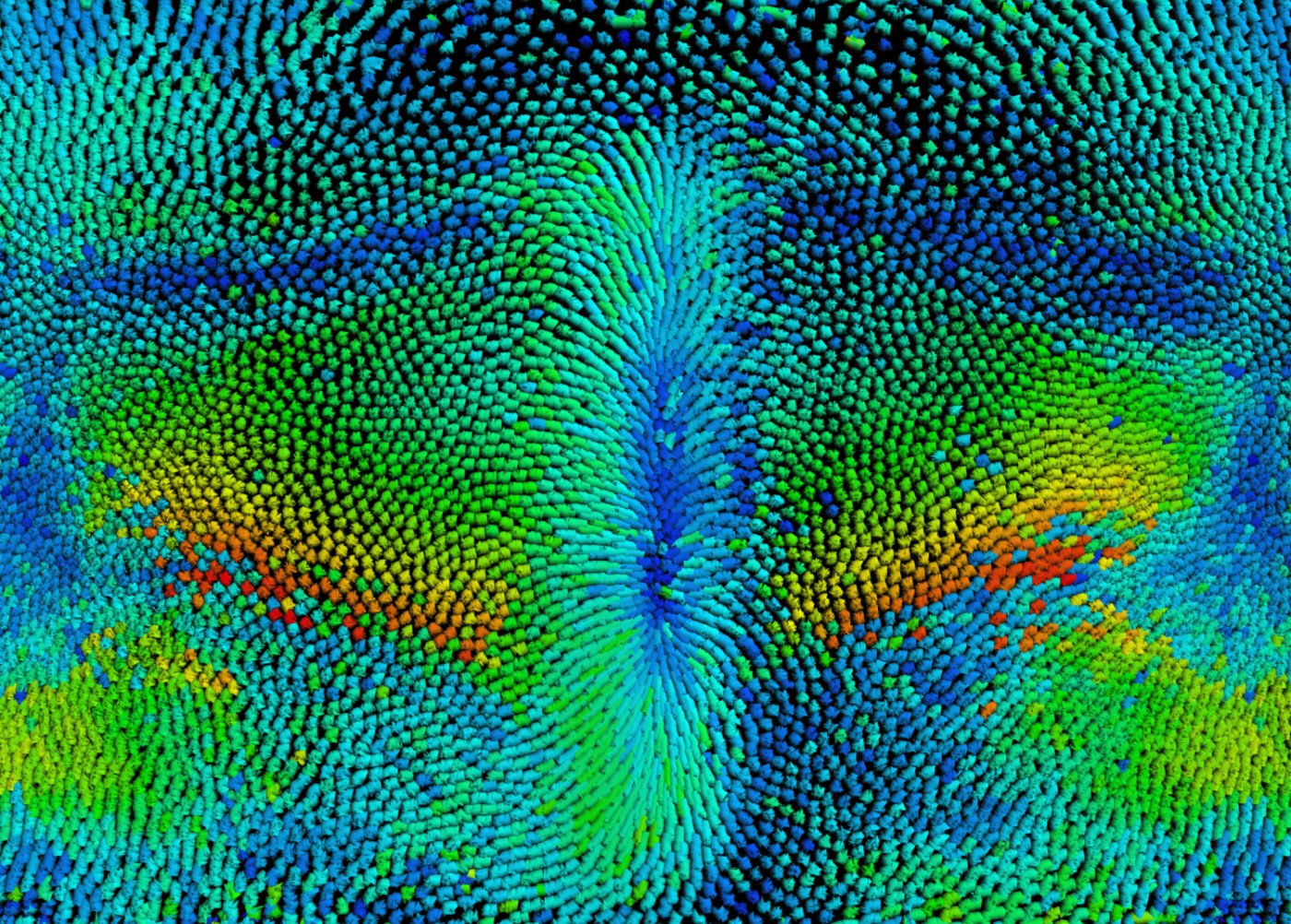
From the molecule to the embryo and back
The research we do in the lab aims to push forward the understanding of tissue morphogenesis and to advance the technology necessary to tackle such understanding. Developing embryos are fascinating and powerful platforms to study the biology and physics of cell collective behavior in a physiological relevant context. In the lab we use and compare two powerful model systems: the protostome Drosophila melanogaster and the deuterostome sea urchin Paracentrotus lividus embryos. While Drosophila provides the most advanced genetic tools, the sea urchin embryo is an ideal system to directly probe cell and tissue mechanics. We implement cutting edge imaging techniques that can provide a comprehensive view of the coordination of tissues at the scale of the embryo with subcellular resolution, laser manipulation, micro-pipetting, optogenetic-based synthetic morphology, big-data processing and multidimensional image analysis. Computational modelling is implemented to delineate a formal physical framework that can theoretically reproduce morphogenetic processes and predict features of the system that are then back tested experimentally. The projects developed in the lab gather people from different backgrounds (biology, computer science, physics and engineering) to generate an interdisciplinary and synergistic group in an international environment.
Figure: cell displacement field shown over a Drosophila embryo cylindrical projection. Red indicates faster cell displacement. Ventral in the center, dorsal on the right and left, anterior top and posterior bottom.
Research Aims
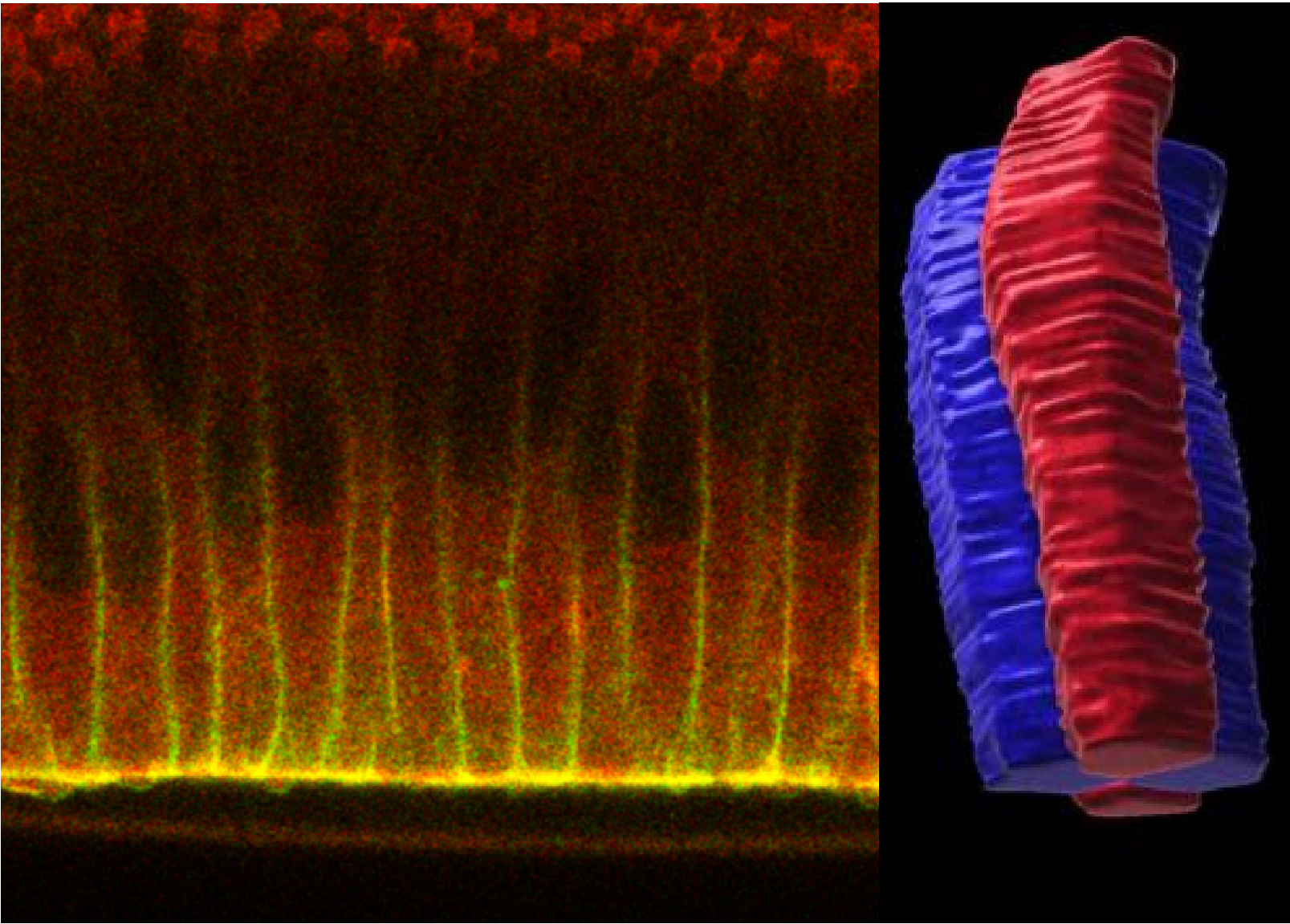
Composite morphogenesis is the process by which a tissue undergoes multiple and simultaneous shape transformations. For instance, during neurulation epithelia extend along the embryo anterior-posterior axis, separating the head from the tail, while simultaneously folding to form the neural tube. How cells can drive multiple and concomitant tissue shape transformations is not known. We have discovered that epithelial cells can devise multiple tiers of adherens junctions with specialized functions at different cell apical-basal positions. We aim to uncover the origin of multi-tier junctions responsible for composite morphogenesis.
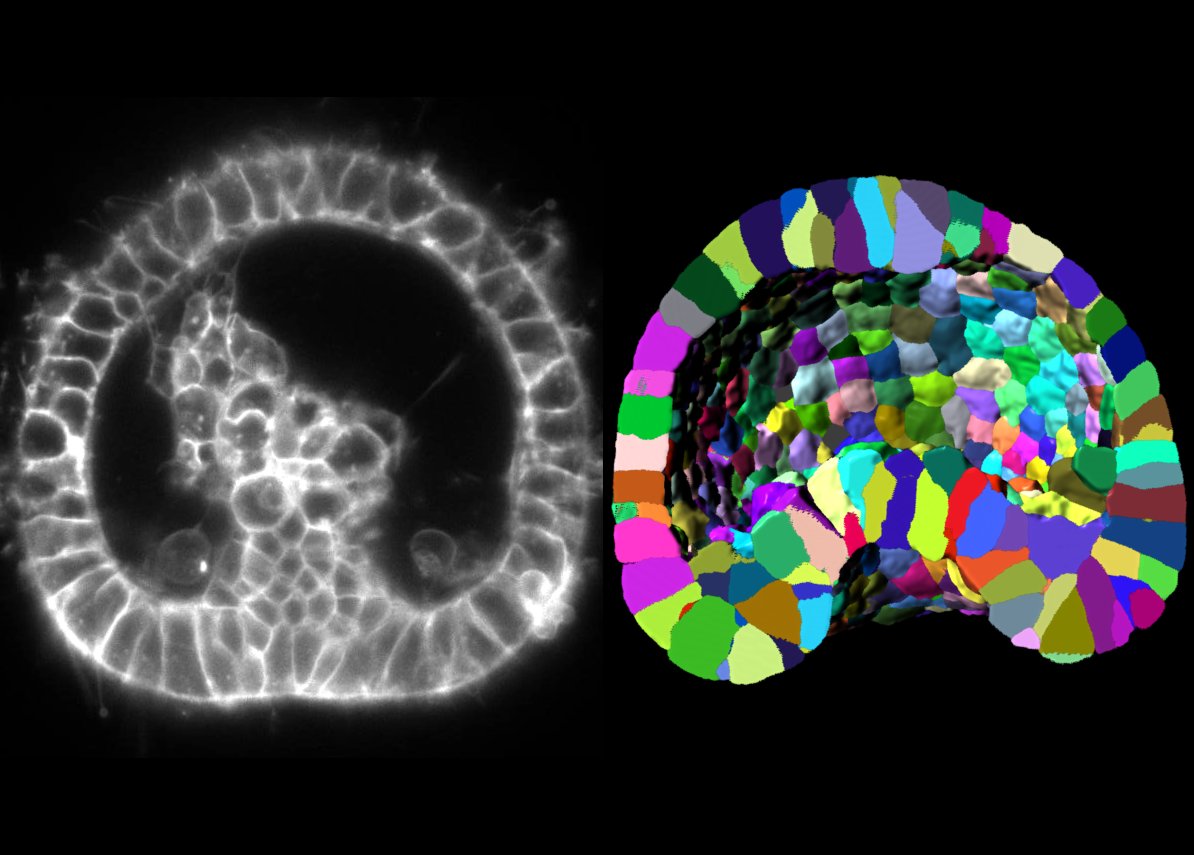
Epithelial tubes play a critical role in multicellular life that is organized in stratified layers and in which an inside and an outside are established. The formation of tubes is essential to build organs responsible to direct vital factors outside-in, inside-out as well as within animals (e.g., food and water through the gut, air through the lungs, electrical signals via the spinal cord, and blood in blood-vessels). Therefore, unveiling the mechanisms responsible for tube formation is key to understand the emergence of complex life forms and to decipher how tubulogenesis disorders, that result from tube formation failure (e.g., spina bifida, polycystic kidney, tracheal atresia), may emerge. To study the formation of epithelial tubes, we use the sea urchin gastrula and focus on the formation of the gut emerging from multiple coordinated and radially planar polarized cell shape changes.
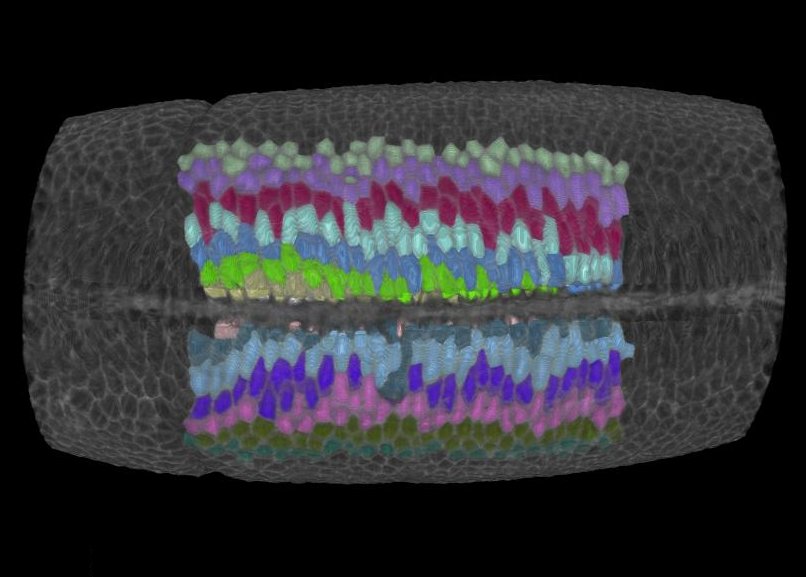
Morphogenetic waves result from the propagation of cell and tissue shape changes across an epithelium. A morphogenetic wave is a shape transformation mode that may be energetically efficient and robust to insure a spatio-temporally continuous tissue shape change resulting in a smooth and coherent structure or organ. Epithelial furrows are often the result of a morphogenetic wave that powers the propagation of a fold along a line resulting in a continuous and linear groove. How fold formation and propagation are initiated, driven and controlled is still poorly understood. To shed new light on the key principles governing morphogenetic waves, we study the molecular mechanisms and the mechanics controlling and driving the formation and the propagation of the ventral and of the cephalic furrow during early Drosophila gastrulation.
Researchers
 DELORME Barthélemy - +33 489150861
DELORME Barthélemy - +33 489150861 POPKOVA Anna - +33 489150861
POPKOVA Anna - +33 489150861
PreDocs
 JALLON Antoine - +33 R
JALLON Antoine - +33 R TANARI Abdul Basith - +33 489150866
TANARI Abdul Basith - +33 489150866 ROBY Nicolas - +33 489150861
ROBY Nicolas - +33 489150861
Engineers & Technicians
 ROUQUET Sami - +33 489150866
ROUQUET Sami - +33 489150866 ROUQUET Sami - +33 489150866
ROUQUET Sami - +33 489150866 AIT MOUFFOK Amel - +33 489150861
AIT MOUFFOK Amel - +33 489150861 GUEYE Mansour - +33 R
GUEYE Mansour - +33 R
Masters
 BEN RHOUMA Elodie - +33 R
BEN RHOUMA Elodie - +33 R
Recent Publications
- Popkova, A, Andrenšek, U, Pagnotta, S, Ziherl, P, Krajnc, M, Rauzi, M et al.. A mechanical wave travels along a genetic guide to drive the formation of an epithelial furrow during Drosophila gastrulation. Dev Cell. 2024;59 (3):400-414.e5. doi: 10.1016/j.devcel.2023.12.016. PubMed PMID:38228140 .
- Fierling, J, John, A, Delorme, B, Torzynski, A, Blanchard, GB, Lye, CM et al.. Embryo-scale epithelial buckling forms a propagating furrow that initiates gastrulation. Nat Commun. 2022;13 (1):3348. doi: 10.1038/s41467-022-30493-3. PubMed PMID:35688832 PubMed Central PMC9187723.
- John, A, Rauzi, M. Composite morphogenesis during embryo development. Semin Cell Dev Biol. 2021;120 :119-132. doi: 10.1016/j.semcdb.2021.06.007. PubMed PMID:34172395 .
- John, A, Rauzi, M. A two-tier junctional mechanism drives simultaneous tissue folding and extension. Dev Cell. 2021;56 (10):1469-1483.e5. doi: 10.1016/j.devcel.2021.04.003. PubMed PMID:33891900 .
- Popkova, A, Rauzi, M, Wang, X. Cellular and Supracellular Planar Polarity: A Multiscale Cue to Elongate the Drosophila Egg Chamber. Front Cell Dev Biol. 2021;9 :645235. doi: 10.3389/fcell.2021.645235. PubMed PMID:33738289 PubMed Central PMC7961075.
- Rauzi, M. Cell intercalation in a simple epithelium. Philos Trans R Soc Lond B Biol Sci. 2020;375 (1809):20190552. doi: 10.1098/rstb.2019.0552. PubMed PMID:32829682 PubMed Central PMC7482223.
- Popkova, A, Stone, OJ, Chen, L, Qin, X, Liu, C, Liu, J et al.. A Cdc42-mediated supracellular network drives polarized forces and Drosophila egg chamber extension. Nat Commun. 2020;11 (1):1921. doi: 10.1038/s41467-020-15593-2. PubMed PMID:32317641 PubMed Central PMC7174421.
- de Medeiros, G, Kromm, D, Balazs, B, Norlin, N, Günther, S, Izquierdo, E et al.. Cell and tissue manipulation with ultrashort infrared laser pulses in light-sheet microscopy. Sci Rep. 2020;10 (1):1942. doi: 10.1038/s41598-019-54349-x. PubMed PMID:32029815 PubMed Central PMC7005178.
- Rauzi, M, Krzic, U, Saunders, TE, Krajnc, M, Ziherl, P, Hufnagel, L et al.. Embryo-scale tissue mechanics during Drosophila gastrulation movements. Nat Commun. 2015;6 :8677. doi: 10.1038/ncomms9677. PubMed PMID:26497898 PubMed Central PMC4846315.
- Collinet, C, Rauzi, M, Lenne, PF, Lecuit, T. Local and tissue-scale forces drive oriented junction growth during tissue extension. Nat Cell Biol. 2015;17 (10):1247-58. doi: 10.1038/ncb3226. PubMed PMID:26389664 .
- Bajoghli, B, Kuri, P, Inoue, D, Aghaallaei, N, Hanelt, M, Thumberger, T et al.. Noninvasive In Toto Imaging of the Thymus Reveals Heterogeneous Migratory Behavior of Developing T Cells. J Immunol. 2015;195 (5):2177-86. doi: 10.4049/jimmunol.1500361. PubMed PMID:26188059 .
- Rauzi, M, Hočevar Brezavšček, A, Ziherl, P, Leptin, M. Physical models of mesoderm invagination in Drosophila embryo. Biophys J. 2013;105 (1):3-10. doi: 10.1016/j.bpj.2013.05.039. PubMed PMID:23823218 PubMed Central PMC3699736.
- Hočevar Brezavšček, A, Rauzi, M, Leptin, M, Ziherl, P. A model of epithelial invagination driven by collective mechanics of identical cells. Biophys J. 2012;103 (5):1069-77. doi: 10.1016/j.bpj.2012.07.018. PubMed PMID:23009857 PubMed Central PMC3433605.
- Rauzi, M, Lenne, PF. Cortical forces in cell shape changes and tissue morphogenesis. Curr Top Dev Biol. 2011;95 :93-144. doi: 10.1016/B978-0-12-385065-2.00004-9. PubMed PMID:21501750 .
- Rauzi, M, Lenne, PF, Lecuit, T. Planar polarized actomyosin contractile flows control epithelial junction remodelling. Nature. 2010;468 (7327):1110-4. doi: 10.1038/nature09566. PubMed PMID:21068726 .
- Bertet, C, Rauzi, M, Lecuit, T. Repression of Wasp by JAK/STAT signalling inhibits medial actomyosin network assembly and apical cell constriction in intercalating epithelial cells. Development. 2009;136 (24):4199-212. doi: 10.1242/dev.040402. PubMed PMID:19934015 .
- Rauzi, M, Lecuit, T. Closing in on mechanisms of tissue morphogenesis. Cell. 2009;137 (7):1183-5. doi: 10.1016/j.cell.2009.06.009. PubMed PMID:19563750 .
- Rauzi, M, Verant, P, Lecuit, T, Lenne, PF. Nature and anisotropy of cortical forces orienting Drosophila tissue morphogenesis. Nat Cell Biol. 2008;10 (12):1401-10. doi: 10.1038/ncb1798. PubMed PMID:18978783 .
- Cavey, M, Rauzi, M, Lenne, PF, Lecuit, T. A two-tiered mechanism for stabilization and immobilization of E-cadherin. Nature. 2008;453 (7196):751-6. doi: 10.1038/nature06953. PubMed PMID:18480755 .
- Rauzi M. Probing tissue interaction with laser-based cauterization in the early developing Drosophila embryo. Methods Cell Biol. 2017;139:153-165. doi: 10.1016/bs.mcb.2016.11.003. Epub 2016 Dec 23. PMID: 28215334.
- Rauzi M, Lenne PF. Probing cell mechanics with subcellular laser dissection of actomyosin networks in the early developing Drosophila embryo. Methods Mol Biol. 2015;1189:209-18. doi: 10.1007/978-1-4939-1164-6_14. PMID: 25245696.
2017 - HFSP CDA
2017 - ATIP-Avenir
2016 - ANR T-ERC
2012 - HFSP Long Term Fellowship
2011 - Embo-Marie Curie Long Term Fellowship

Do not miss an exiting talk by Sylvain Gabriele from University of Mons on epithelia sensing curvature and spatial confinement
Read More

Do not miss an exciting talk accessible to an interdisciplinary audience on how aquatic creatures optimize their swimming
Read More

Popkova et al. from the Rauzi team publish in Dev. Cell a pioneer work on morphogenetic waves during embryonic development.
Read More

MECABIONIC Symposium – Mechanobiology across scales: from the molecule to the organism and back
Read More
The embryo puts on a string to initiate gastrulation
Read More
A two-tier junctional mechanism drives composite morphogenesis
Read More
The egg puts on a corset to get the right shape
Read More
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2



