
Christian BRAENDLE
Gene-environment interactions in development and evolution
Main interests
- Phenotypic plasticity, genotype-by-environment interactions and genetic assimilation
- Genotype-phenotype map: from developmental variation to life history variation
- Evolution and ecology of Caenorhabditis nematodes
Scientific Questions
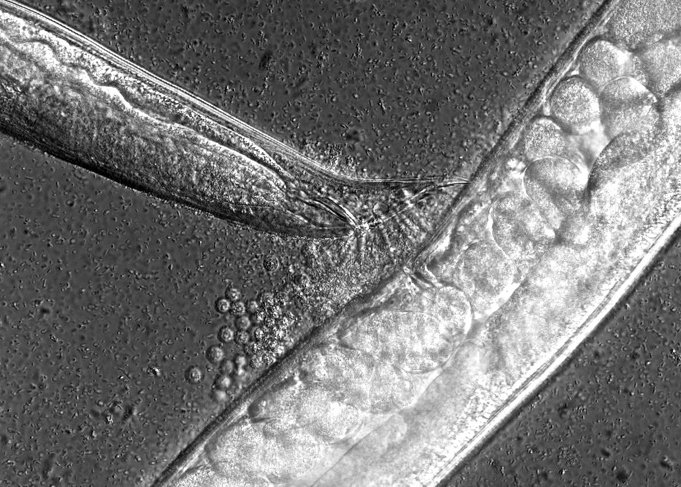
Environmental variation is a key force shaping the function and evolution of developmental mechanisms. Therefore, it is essential to understand how environmental variation modulates developmental processes and resulting phenotypic variation. Much of our research therefore addresses the questions of how developmental systems respond to environmental variation and how such responses evolve. Using the nematode Caenorhabditis elegans and related species, we aim to answer four main questions: How does development integrate variable environmental information to alter phenotypic outcomes? How do developmental responses to environmental variation evolve? How do quantiative developmental traits evolve within species? What are developmental changes underlying adaptive evolution of life histories? We investigate these questions by focusing on molecularly well-characterized processes, such as vulval cell fate patterning, germline development and dauer formation.
Our Strategy
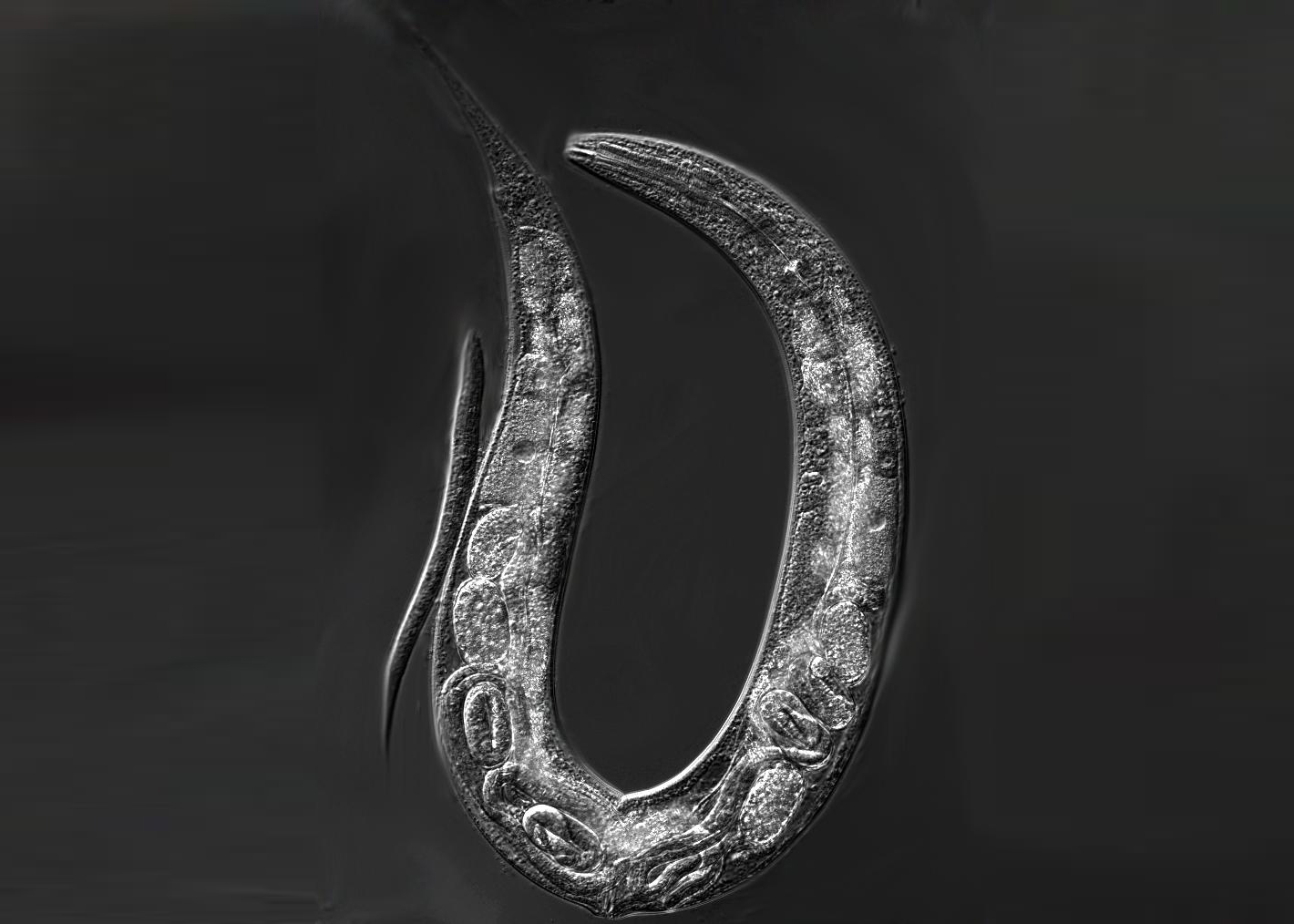
Although genotype-by-environment interactions are common and important determinants of phenotypic variation, the mechanisms by which genetic and environmental variation interact to generate trait variation remain poorly understood. Our projects thus aim to characterize the molecular and developmental basis of genotype-by-environment interactions, how such interactions evolve and how they in turn may impact the evolutionary process itself. In our research, we use the nematode Caenorhabditis elegans and related species as model organisms, and we integrate quantitative experimental approaches from developmental and evolutionary genetics.
Research Aims
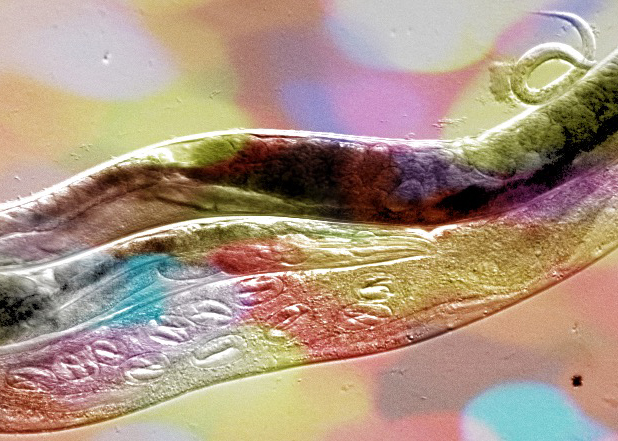
Phenotypic plasticity, genotype-by-environment interactions and genetic assimilation
We focus on the environmental context-dependence of developmental processes (e.g. germ cell proliferation, gametogenesis, dauer formation) to characterize the molecular basis and evolution of phenotypic plasticity. Primarily, we aim to identify developmental and molecular determinants of natural variation in these phenotypes through a combination of quantitative and developmental genetic approaches.
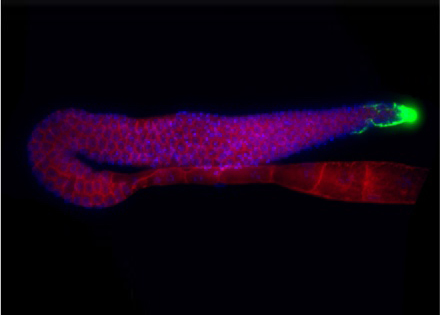
Genotype-phenotype map: from developmental variation to life history variation
How complex traits, such as life history traits, emerge through developmental integration of numerous gene-environment, gene-gene interactions, and higher-order interactions among cells and tissues, remains largely unknown. We therefore study the nematode hermaphrodite germline to ask how variation in different parameters of this developmental system translates into variation in reproductive life histories.
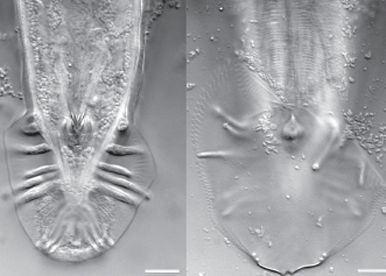
Evolution and ecology of Caenorhabditis nematodes
Although C. elegans is increasingly being used in evolutionary studies, there is still very little information on its natural history, ecology, phylogenetic context, and the genetic structure of its natural populations. We therefore contribute to current community efforts to sample and characterize natural Caenorhabditis populations to generate a more comprehensive evolutionary ecological context for C. elegans and its close relatives.
Researchers
 GIMOND Clotilde - +33 489150842
GIMOND Clotilde - +33 489150842 MIGNEROT-FUSTE Laure - +33 489150842
MIGNEROT-FUSTE Laure - +33 489150842
Postdocs
 GONZALEZ SOMERMEYER Louisa - +33 R
GONZALEZ SOMERMEYER Louisa - +33 R
PreDocs
 LE PARC Amélie - +33 489150842
LE PARC Amélie - +33 489150842
Engineers & Technicians
 SANDJAK Asma - +33 489150842
SANDJAK Asma - +33 489150842 BOUVET Orianne - +33 489150842
BOUVET Orianne - +33 489150842 LEROUX Delphine - +33 R
LEROUX Delphine - +33 R BOYER Salomé - +33 R
BOYER Salomé - +33 R
Students
 GOMES SANCHES VAZ Eliane - +33 R
GOMES SANCHES VAZ Eliane - +33 R
Recent publications
- Braendle, C, Paaby, A. Life history in Caenorhabditis elegans: from molecular genetics to evolutionary ecology. Genetics. 2024;228 (3):. doi: 10.1093/genetics/iyae151. PubMed PMID:39422376 PubMed Central PMC11538407.
- Mignerot, L, Gimond, C, Bolelli, L, Bouleau, C, Sandjak, A, Boulin, T et al.. Natural variation in the Caenorhabditis elegans egg-laying circuit modulates an intergenerational fitness trade-off. Elife. 2024;12 :. doi: 10.7554/eLife.88253. PubMed PMID:38564369 PubMed Central PMC10987095.
- Fausett, SR, Laury, CA, Magallon, RE, Braendle, C. Simplified Quantification of Progenitor Zone Size, an Indicator of Germ Stem Cell Niche Activity, in the Nematode Caenorhabditis elegans. Methods Mol Biol. 2024; :. doi: 10.1007/7651_2024_536. PubMed PMID:38507211 .
- Fausett, SR, Sandjak, A, Billard, B, Braendle, C. Higher-order epistasis shapes natural variation in germ stem cell niche activity. Nat Commun. 2023;14 (1):2824. doi: 10.1038/s41467-023-38527-0. PubMed PMID:37198172 PubMed Central PMC10192456.
- Huang, Y, Lo, YH, Hsu, JC, Le, TS, Yang, FJ, Chang, T et al.. Widespread sex ratio polymorphism in Caenorhabditis nematodes. R Soc Open Sci. 2023;10 (3):221636. doi: 10.1098/rsos.221636. PubMed PMID:36938539 PubMed Central PMC10014251.
- Gimond, C, Poullet, N, Braendle, C. Isolating Caenorhabditis elegans from the Natural Habitat. Methods Mol Biol. 2022;2468 :283-292. doi: 10.1007/978-1-0716-2181-3_15. PubMed PMID:35320571 .
- Fausett, S, Poullet, N, Gimond, C, Vielle, A, Bellone, M, Braendle, C et al.. Germ cell apoptosis is critical to maintain Caenorhabditis elegans offspring viability in stressful environments. PLoS One. 2021;16 (12):e0260573. doi: 10.1371/journal.pone.0260573. PubMed PMID:34879088 PubMed Central PMC8654231.
- Grover, M, Fasseas, MK, Essmann, C, Liu, K, Braendle, C, Félix, MA et al.. Infection of C. elegans by Haptoglossa Species Reveals Shared Features in the Host Response to Oomycete Detection. Front Cell Infect Microbiol. 2021;11 :733094. doi: 10.3389/fcimb.2021.733094. PubMed PMID:34722333 PubMed Central PMC8552708.
- Lee, D, Zdraljevic, S, Stevens, L, Wang, Y, Tanny, RE, Crombie, TA et al.. Balancing selection maintains hyper-divergent haplotypes in Caenorhabditis elegans. Nat Ecol Evol. 2021;5 (6):794-807. doi: 10.1038/s41559-021-01435-x. PubMed PMID:33820969 PubMed Central PMC8202730.
- Vigne, P, Gimond, C, Ferrari, C, Vielle, A, Hallin, J, Pino-Querido, A et al.. A single-nucleotide change underlies the genetic assimilation of a plastic trait. Sci Adv. 2021;7 (6):. doi: 10.1126/sciadv.abd9941. PubMed PMID:33536214 PubMed Central PMC7857674.
- Noble, LM, Yuen, J, Stevens, L, Moya, N, Persaud, R, Moscatelli, M et al.. Selfing is the safest sex for Caenorhabditis tropicalis. Elife. 2021;10 :. doi: 10.7554/eLife.62587. PubMed PMID:33427200 PubMed Central PMC7853720.
- Ben-David, E, Pliota, P, Widen, SA, Koreshova, A, Lemus-Vergara, T, Verpukhovskiy, P et al.. Ubiquitous Selfish Toxin-Antidote Elements in Caenorhabditis Species. Curr Biol. 2021;31 (5):990-1001.e5. doi: 10.1016/j.cub.2020.12.013. PubMed PMID:33417886 .
- Billard, B, Vigne, P, Braendle, C. A Natural Mutational Event Uncovers a Life History Trade-Off via Hormonal Pleiotropy. Curr Biol. 2020;30 (21):4142-4154.e9. doi: 10.1016/j.cub.2020.08.004. PubMed PMID:32888477 .
- Billard, B, Gimond, C, Braendle, C. [Genetics and evolution of developmental plasticity in the nematode C. elegans: Environmental induction of the dauer stage]. Biol Aujourdhui. 2020;214 (1-2):45-53. doi: 10.1051/jbio/2020006. PubMed PMID:32773029 .
- Lee, D, Zdraljevic, S, Cook, DE, Frézal, L, Hsu, JC, Sterken, MG et al.. Selection and gene flow shape niche-associated variation in pheromone response. Nat Ecol Evol. 2019;3 (10):1455-1463. doi: 10.1038/s41559-019-0982-3. PubMed PMID:31548647 PubMed Central PMC6764921.
- Gimond, C, Vielle, A, Silva-Soares, N, Zdraljevic, S, McGrath, PT, Andersen, EC et al.. Natural Variation and Genetic Determinants of Caenorhabditis elegans Sperm Size. Genetics. 2019;213 (2):615-632. doi: 10.1534/genetics.119.302462. PubMed PMID:31395653 PubMed Central PMC6781899.
- Shin, H, Braendle, C, Monahan, KB, Kaplan, REW, Zand, TP, Mote, FS et al.. Developmental fidelity is imposed by genetically separable RalGEF activities that mediate opposing signals. PLoS Genet. 2019;15 (5):e1008056. doi: 10.1371/journal.pgen.1008056. PubMed PMID:31086367 PubMed Central PMC6534338.
- Stevens, L, Félix, MA, Beltran, T, Braendle, C, Caurcel, C, Fausett, S et al.. Comparative genomics of 10 new Caenorhabditis species. Evol Lett. 2019;3 (2):217-236. doi: 10.1002/evl3.110. PubMed PMID:31007946 PubMed Central PMC6457397.
- Frézal, L, Demoinet, E, Braendle, C, Miska, E, Félix, MA. Natural Genetic Variation in a Multigenerational Phenotype in C. elegans. Curr Biol. 2018;28 (16):2588-2596.e8. doi: 10.1016/j.cub.2018.05.091. PubMed PMID:30078564 PubMed Central PMC6984962.
- Grimbert, S, Vargas Velazquez, AM, Braendle, C. Physiological Starvation Promotes Caenorhabditis elegans Vulval Induction. G3 (Bethesda). 2018;8 (9):3069-3081. doi: 10.1534/g3.118.200449. PubMed PMID:30037804 PubMed Central PMC6118308.
2012 - Fellow at the Institute for Advanced Study, Berlin
2010 - Schlumberger Award
2008 - ATIP, CNRS
1995 - Fund for Young Talented People, Swiss Study Foundation
1994 - Winner of highest award in the Swiss science contest for young people “Schweizer Jugend Forscht”

Christian Braendle awarded HFSP research grant for groundbreaking work on adaptation to extreme environments
Read More
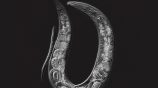
Maternal self-sacrifice in C. elegans: an example of genetic assimilation
Read More
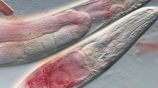
Evolution of a trade-off between two life history traits in the nematode C. elegans.
Read More

When iBV researchers meet Art at the Museum of Modern Art
Read More

Genetic basis of sperm size in C. elegans: a role for the chromatin remodeling complex NURF-1
Read More
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
