
Thierry VIROLLE
Cancer Stem Cell Plasticity and Functional Intra-tumor Heterogeneity
Main interests
- Brain Tumor
- Cancer stem cells
- Intra-tumor heterogeneity
- Epigenetic regulation
Scientific Questions
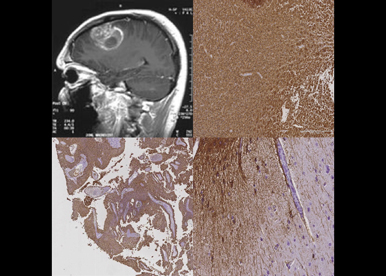
Glioblastomas (GBM) are the most common form of primary brain tumors in adult. These tumors remain incurable and lead to a fatal outcome in less than 18 months. Tumor development is a complex process mixing normal cells and heterogeneous cancer cell populations. A sublevel of complexity in intra-tumor heterogeneity resides in the co-existence of functionally divergent heterogeneous populations of cells. GBM exemplifies this situation by harboring tumor territories functionally divergent while sharing similar genomic abnormalities. These territories are enriched in mitotic tumor cells, positive for markers of GBM stem cell (GSC), or enriched in non-mitotic, differentiated cells, frequently associated to pathological vessels and large necrotic areas. Such heterogeneity constitutes a major obstacle for conventional therapies, mostly due to the selection and expansion of resistant GSC. Thus targeting GSC properties and mitotic territories will be valuable for therapeutic strategies.
Our Strategy
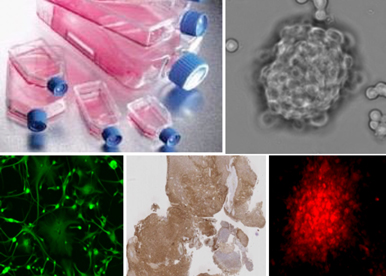
The research developed by our group focuses on the study of the regulation of GSC plasticity in the context of the genesis of tumor micro-territories. Through a better understanding of tumor homeostasis, our goal is to identify key molecular factors of interest for therapeutic purposes. The group is composed of researchers and clinicians, an essential association to link fundamental research with clinical applications. Because GBM are heterogeneous tumors, we have built a platform dedicated to establish and characterize a collection of GSC primary cultures (from different patients) as well as GBM samples representative of mitotic and non-mitotic tumor territories. This collection, crucial for our research, allowed us to identify key molecular factors involved in the control of GSC properties and tumorigenicity.
The objective of our research development is to model the molecular basis involved in the biogenesis and homeostasis of both mitotic and non-mitotic tumor territories, affecting tumor progression and aggressiveness. Our aim is to identify innovative therapeutic targets capable to efficiently repress GSC properties.
We have identified a tumor suppressor miRNA signature (among them the miR-302-367 cluster), constituting a feature and a gatekeeper of non-mitotic territories, while one of its common targets, a splice factor, is restricted to stem-like/progenitor cells in mitotic territories and contributes to the maintenance of stemness and selfrenewal.
Research Aims
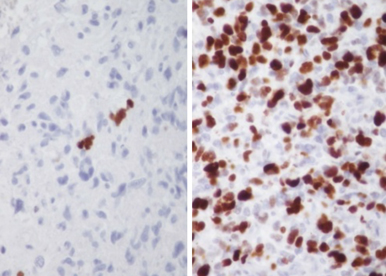
We will characterize the regulation of this splice factor and its mode of action in self-renewing GSC by identifying its most functionally prominent targets (deep RNAseq, exome analysis and CLIP-seq). Our recent results indicate the strong contribution of this factor in the regulation of H3K9 and H3K27 methylation, influencing thus the chromatin structure. Using bioinformatic and mathematical modeling, we will determine hubs of regulatory targets involved in GSC maintenance and aggressiveness.
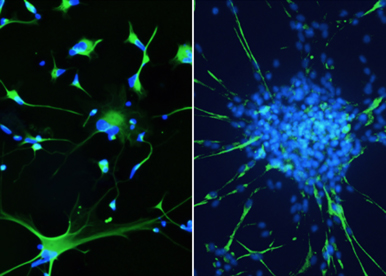
Signals coming from the tumor microenvironment including soluble proteins, extracellular matrix components, secreted vesicles as well as tunneling nanotubes (TNT) are most likely important factors to guide GBM cell fate decision. This part of our project is to functionally characterize these cell-to-cell communication processes and verify their contribution in the genesis of functionally divergent tumor territories.
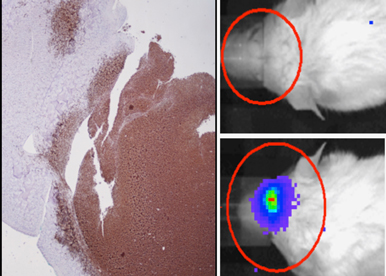
The development of molecules capable in promoting irreversible loss of CSC populations, through the modulation of the specific factors that we identified (miRNA, transcription factors, splice factors), will constitute an innovative and attractive therapeutic approach compatible with industrial pipelines. We develop in tight collaborations with chemists (Dr M Duca, ICN Nice; Dr F Roussy, Dr M Litaudon, ICSN Gyf sur Yvette) the characterization of innovative compounds targeting GSC.
Researchers
 LADOUX Annie - +33 489150821
LADOUX Annie - +33 489150821 LADOUX Annie - +33 489150821
LADOUX Annie - +33 489150821 SALIOU Alexa - +33 R
SALIOU Alexa - +33 R RABY Amélie - +33 R
RABY Amélie - +33 R BOULAKIRBA Sonia - +33 R
BOULAKIRBA Sonia - +33 R
Clinical Researchers
 PAQUIS Philippe - +33 489150821
PAQUIS Philippe - +33 489150821 BUREL VANDENBOS Fanny - +33 489150821
BUREL VANDENBOS Fanny - +33 489150821 ALMAIRAC Fabien - +33 489150821
ALMAIRAC Fabien - +33 489150821
Engineers & Technicians
 POLO Béatrice - +33 489150821
POLO Béatrice - +33 489150821 TURCHI Laurent - +33 489150820
TURCHI Laurent - +33 489150820 PEUX Clément - +33 489150821
PEUX Clément - +33 489150821
Students
 PERON Camille - +33 R
PERON Camille - +33 R
- Turchi, L, Sakakini, N, Saviane, G, Polo, B, Saurty-Seerunghen, MS, Gabut, M et al.. Correction: Turchi et al. CELF2 Sustains a Proliferating/OLIG2+ Glioblastoma Cell Phenotype via the Epigenetic Repression of SOX3. Cancers 2023, 15, 5038. Cancers (Basel). 2025;17 (9):. doi: 10.3390/cancers17091583. PubMed PMID:40361521 PubMed Central PMC12072141.
- Narayanan, A, Saurty-Seerunghen, MS, Michieletto, J, Delaunay, V, Bruneel, A, Dupré, T et al.. Nicotinamide metabolism reprogramming drives reversible senescence of glioblastoma cells. Cell Mol Life Sci. 2025;82 (1):126. doi: 10.1007/s00018-025-05641-9. PubMed PMID:40116940 PubMed Central PMC11928343.
- Chardin, D, Jing, L, Chazal-Ngo-Mai, M, Guigonis, JM, Rigau, V, Goze, C et al.. Identification of Metabolomic Markers in Frozen or Formalin-Fixed and Paraffin-Embedded Samples of Diffuse Glioma from Adults. Int J Mol Sci. 2023;24 (23):. doi: 10.3390/ijms242316697. PubMed PMID:38069019 PubMed Central PMC10705927.
- Turchi, L, Sakakini, N, Saviane, G, Polo, B, Saurty-Seerunghen, MS, Gabut, M et al.. CELF2 Sustains a Proliferating/OLIG2+ Glioblastoma Cell Phenotype via the Epigenetic Repression of SOX3. Cancers (Basel). 2023;15 (20):. doi: 10.3390/cancers15205038. PubMed PMID:37894405 PubMed Central PMC10605641.
- Jézéquel, G, Rampal, C, Guimard, C, Kovacs, D, Polidori, J, Bigay, J et al.. Structure-Based Design of a Lead Compound Derived from Natural Schweinfurthins with Antitumor Properties That Target Oxysterol-Binding Protein. J Med Chem. 2023;66 (20):14208-14220. doi: 10.1021/acs.jmedchem.3c01298. PubMed PMID:37795600 .
- Nakhle, J, Khattar, K, Özkan, T, Boughlita, A, Abba Moussa, D, Darlix, A et al.. Mitochondria Transfer from Mesenchymal Stem Cells Confers Chemoresistance to Glioblastoma Stem Cells through Metabolic Rewiring. Cancer Res Commun. 2023;3 (6):1041-1056. doi: 10.1158/2767-9764.CRC-23-0144. PubMed PMID:37377608 PubMed Central PMC10266428.
- Bikfalvi, A, da Costa, CA, Avril, T, Barnier, JV, Bauchet, L, Brisson, L et al.. Challenges in glioblastoma research: focus on the tumor microenvironment: (Trends in Cancer, 9:1 p:9-27, 2023). Trends Cancer. 2023;9 (8):692. doi: 10.1016/j.trecan.2023.02.006. PubMed PMID:36997420 .
- Bikfalvi, A, da Costa, CA, Avril, T, Barnier, JV, Bauchet, L, Brisson, L et al.. Challenges in glioblastoma research: focus on the tumor microenvironment. Trends Cancer. 2023;9 (1):9-27. doi: 10.1016/j.trecan.2022.09.005. PubMed PMID:36400694 .
- Saurty-Seerunghen, MS, Daubon, T, Bellenger, L, Delaunay, V, Castro, G, Guyon, J et al.. Glioblastoma cell motility depends on enhanced oxidative stress coupled with mobilization of a sulfurtransferase. Cell Death Dis. 2022;13 (10):913. doi: 10.1038/s41419-022-05358-8. PubMed PMID:36310164 PubMed Central PMC9618559.
- Lié, O, Virolle, T, Gabut, M, Pasquier, C, Zemmoura, I, Augé-Gouillou, C et al.. SETMAR Shorter Isoform: A New Prognostic Factor in Glioblastoma. Front Oncol. 2021;11 :638397. doi: 10.3389/fonc.2021.638397. PubMed PMID:35047379 PubMed Central PMC8761672.
- Almairac, F, Turchi, L, Sakakini, N, Debruyne, DN, Elkeurti, S, Gjernes, E et al.. ERK-Mediated Loss of miR-199a-3p and Induction of EGR1 Act as a "Toggle Switch" of GBM Cell Dedifferentiation into NANOG- and OCT4-Positive Cells. Cancer Res. 2020;80 (16):3236-3250. doi: 10.1158/0008-5472.CAN-19-0855. PubMed PMID:32366479 .
- Torres, Á, Erices, JI, Sanchez, F, Ehrenfeld, P, Turchi, L, Virolle, T et al.. Extracellular adenosine promotes cell migration/invasion of Glioblastoma Stem-like Cells through A3 Adenosine Receptor activation under hypoxia. Cancer Lett. 2019;446 :112-122. doi: 10.1016/j.canlet.2019.01.004. PubMed PMID:30660649 .
- Virolle, T. [Cancer stem cells in glioblastoma]. Bull Cancer. 2017;104 (12):1075-1079. doi: 10.1016/j.bulcan.2017.10.012. PubMed PMID:29153545 .
- Bogeas, A, Morvan-Dubois, G, El-Habr, EA, Lejeune, FX, Defrance, M, Narayanan, A et al.. Changes in chromatin state reveal ARNT2 at a node of a tumorigenic transcription factor signature driving glioblastoma cell aggressiveness. Acta Neuropathol. 2018;135 (2):267-283. doi: 10.1007/s00401-017-1783-x. PubMed PMID:29149419 PubMed Central PMC5773658.
- Dadone-Montaudié, B, Ambrosetti, D, Dufour, M, Darcourt, J, Almairac, F, Coyne, J et al.. [18F] FDOPA standardized uptake values of brain tumors are not exclusively dependent on LAT1 expression. PLoS One. 2017;12 (9):e0184625. doi: 10.1371/journal.pone.0184625. PubMed PMID:28937983 PubMed Central PMC5609741.
- Debruyne, DN, Turchi, L, Burel-Vandenbos, F, Fareh, M, Almairac, F, Virolle, V et al.. DOCK4 promotes loss of proliferation in glioblastoma progenitor cells through nuclear beta-catenin accumulation and subsequent miR-302-367 cluster expression. Oncogene. 2018;37 (2):241-254. doi: 10.1038/onc.2017.323. PubMed PMID:28925399 .
- Fareh, M, Almairac, F, Turchi, L, Burel-Vandenbos, F, Paquis, P, Fontaine, D et al.. Cell-based therapy using miR-302-367 expressing cells represses glioblastoma growth. Cell Death Dis. 2017;8 (3):e2713. doi: 10.1038/cddis.2017.117. PubMed PMID:28358371 PubMed Central PMC5386523.
- El-Habr, EA, Dubois, LG, Burel-Vandenbos, F, Bogeas, A, Lipecka, J, Turchi, L et al.. A driver role for GABA metabolism in controlling stem and proliferative cell state through GHB production in glioma. Acta Neuropathol. 2017;133 (4):645-660. doi: 10.1007/s00401-016-1659-5. PubMed PMID:28032215 PubMed Central PMC5348560.
- Papin-Michault, C, Bonnetaud, C, Dufour, M, Almairac, F, Coutts, M, Patouraux, S et al.. Study of LAT1 Expression in Brain Metastases: Towards a Better Understanding of the Results of Positron Emission Tomography Using Amino Acid Tracers. PLoS One. 2016;11 (6):e0157139. doi: 10.1371/journal.pone.0157139. PubMed PMID:27276226 PubMed Central PMC4898730.
- Sakakini, N, Turchi, L, Bergon, A, Holota, H, Rekima, S, Lopez, F et al.. A Positive Feed-forward Loop Associating EGR1 and PDGFA Promotes Proliferation and Self-renewal in Glioblastoma Stem Cells. J Biol Chem. 2016;291 (20):10684-99. doi: 10.1074/jbc.M116.720698. PubMed PMID:27002148 PubMed Central PMC4865916.
2014 - Prix de l'appel à projet santé du conseil général 06 - Conseil départemental
1998 - prix Coloplast - entreprise Coloplast

Thierry Virolle’s team highlighted in Nice Matin and La Tribune for the creation of a new start up, Virtu Therapeutics, aimed to develop innovative therapeutic strategies to fight against glioblastoma!
Read More
iBV Researchers and clinicians join forces to fight glioblastoma
Read More
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
