
Arnaud HUBSTENBERGER
Epitranscriptomics
Main interests
- RNA condensates, phase separations, phase transitions, and the multiscale structure of the transcriptome
- From single genes to the expression of coherent networks: how RNA supramolecular co-assemblies allow RNAs to coordinately fine tune gene expression
- Robustness, noise and plasticity of RNA networks and regulons: The synchronization of RNA expression to development, and its adaptation to environmental changes
- Epitranscriptomics: how RNPs are marked by cellular history
Scientific Questions
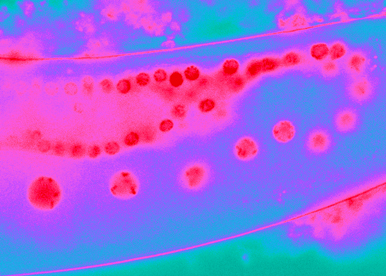
Gene networks must be synchronized to development, adapted to the environment, and marked by cell lineage and past history.
The genome can be compared to a dictionary of thousands of individual genes that can be transcribed into RNA, vehicle of the genetic information. In the same manner that words need to be pronounced at the right time in the right place to generate sentences that carry a coherent message, RNA molecules must be coordinately expressed. The molecular grammar that organize RNA within cells remain poorly understood. As a part of this process, RNAs are covered by proteins that will regulate RNA storage, decay, expression, repression, and localization. Multiple RNAs can further co-assemble into condensates. In this context, we focus on the posttranscriptional control of gene expression:
1-What is the multiscale and multiphase organization of the transcriptome?
2-How does it coordinate RNA expression during development?
3-Adapt it to the environment, stress and aging?
Our Strategy
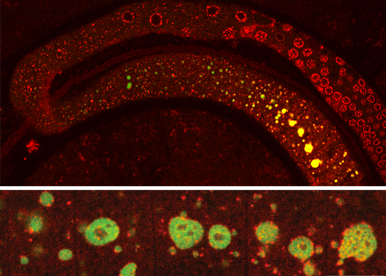
We do not consider RNA as isolated single molecules that belong to an homogeneous soluble phase within the cytosol. Soluble RNA can diffuse, condense into liquid droplets or viscoelastic hydrogels, or even freeze into disorganized glass like aggregates or quasi-crystalline solids. We and others have introduced phase transition as a means to study RNA organization, regulation, and function.
Our hypothesis is that RNA condensates structure and mark the RNA information. The resulting organization is regulated by developmental cues, environmental cues, and cellular history: phase separation participates in the collective control of RNA expression.
Our strategy is to model how phase separations structure the cytosolic transcriptome, and test its functionality in coordinating RNA expression. As models, we use C elegans germline development and human cells in culture.
(1) To study the structural principles of RNA supramolecular organization, a FAPS method to purify diverse RNA condensates, and separate various RNA phases, is coupled to transcriptomics and proteomics.
(2) To model the transcriptome 3D organization and structural fluidity we map RNA-RNA inter and intra molecular interactions, adapting high throughput sequencing methods.
(3) To integrate single molecule behavior to global RNA regulation through phase separation, we develop new imaging assay for RNA localization and translation as well as methods to probe the material emerging properties of RNA condensates.
Research Aims
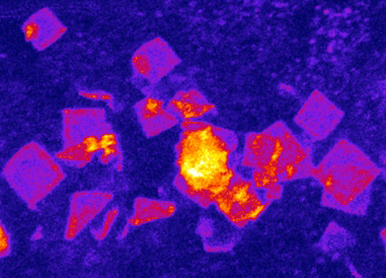
MAPING THE DIVERSITY AND MODELING THE STRUCTURE OF RNP CONDENSATES
To model the multiphase and multiscale organization of the transcriptome in the cytosol, we will further develop new high-throughput approaches that (1) identify RNAs and proteins enriched in diverse RNP condensates, (2) map short/long range RNA-RNA and RNA-protein interactions, and (3) distinguish the soluble/condensed, liquid/solid, structured/disorganized states of RNAs.
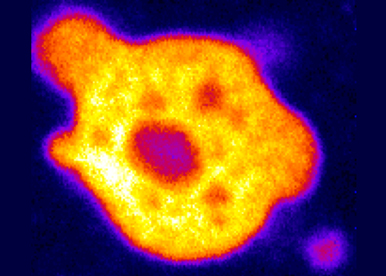
TESTING THE FUNCTION OF RNP CONDENSATES IN CORDINATING RNA REGULONS
At steady state, during developmental switches, and in response to the environment, we will test how RNP condensates and phase transitions sort, buffer, tune and coordinate RNA regulons. For that purpose, we will genetically manipulate condensates and combine single molecule imaging with transcriptomic approaches.
Researchers
 CIAIS Delphine - +33 489150791
CIAIS Delphine - +33 489150791
PreDocs
 OUERTANI Sarah - +33 489150865
OUERTANI Sarah - +33 489150865 BAHRI Alia - +33 489150866
BAHRI Alia - +33 489150866 VALéRO Florian - +33 489150866
VALéRO Florian - +33 489150866 LERAY Chloé - +33 R
LERAY Chloé - +33 R LAGHRISSI Hiba - +33 489150746
LAGHRISSI Hiba - +33 489150746 VAYANKARA EDACHOLA Sreeparvathy - +33 489150878
VAYANKARA EDACHOLA Sreeparvathy - +33 489150878
Engineers & Technicians
 BRUNI-FAVIER Solène - +33 R
BRUNI-FAVIER Solène - +33 R JACQUET Karine - +33 R
JACQUET Karine - +33 R
Masters
 HOTA Prangya - +33 R
HOTA Prangya - +33 R
Recent publications
- Cardona, AH, Ecsedi, S, Khier, M, Yi, Z, Bahri, A, Ouertani, A et al.. Self-demixing of mRNA copies buffers mRNA:mRNA and mRNA:regulator stoichiometries. Cell. 2023;186 (20):4310-4324.e23. doi: 10.1016/j.cell.2023.08.018. PubMed PMID:37703874 .
- Vidal-Cruchez, O, Nicolini, VJ, Rete, T, Jacquet, K, Rezzonico, R, Lacoux, C et al.. KRAS and NRAS Translation Is Increased upon MEK Inhibitors-Induced Processing Bodies Dissolution. Cancers (Basel). 2023;15 (12):. doi: 10.3390/cancers15123078. PubMed PMID:37370689 PubMed Central PMC10296394.
- Adekunle, DA, Hubstenberger, A. The multiscale and multiphase organization of the transcriptome. Emerg Top Life Sci. 2020;4 (3):265-280. doi: 10.1042/ETLS20190187. PubMed PMID:32542380 .
- Courel, M, Clément, Y, Bossevain, C, Foretek, D, Vidal Cruchez, O, Yi, Z et al.. GC content shapes mRNA storage and decay in human cells. Elife. 2019;8 :. doi: 10.7554/eLife.49708. PubMed PMID:31855182 PubMed Central PMC6944446.
- De Graeve, F, Debreuve, E, Rahmoun, S, Ecsedi, S, Bahri, A, Hubstenberger, A et al.. Detecting and quantifying stress granules in tissues of multicellular organisms with the Obj.MPP analysis tool. Traffic. 2019;20 (9):697-711. doi: 10.1111/tra.12678. PubMed PMID:31314165 .
- Courel, M, Bénard, M, Ernoult-Lange, M, Chouaib, R, Hubstenberger, A, Kress, M et al.. [P-bodies: microscopic droplets to store mRNAs encoding regulatory proteins]. Med Sci (Paris). 2018;34 (4):306-308. doi: 10.1051/medsci/20183404009. PubMed PMID:29658471 .
- Hubstenberger, A, Courel, M, Bénard, M, Souquere, S, Ernoult-Lange, M, Chouaib, R et al.. P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell. 2017;68 (1):144-157.e5. doi: 10.1016/j.molcel.2017.09.003. PubMed PMID:28965817 .
- Hubstenberger, A, Cameron, C, Noble, SL, Keenan, S, Evans, TC. Modifiers of solid RNP granules control normal RNP dynamics and mRNA activity in early development. J Cell Biol. 2015;211 (3):703-16. doi: 10.1083/jcb.201504044. PubMed PMID:26527741 PubMed Central PMC4639854.
- Li, S, Lamarche, F, Charton, R, Delphin, C, Gires, O, Hubstenberger, A et al.. Expression analysis of ATAD3 isoforms in rodent and human cell lines and tissues. Gene. 2014;535 (1):60-9. doi: 10.1016/j.gene.2013.10.062. PubMed PMID:24239551 .
- Hubstenberger, A, Noble, SL, Cameron, C, Evans, TC. Translation repressors, an RNA helicase, and developmental cues control RNP phase transitions during early development. Dev Cell. 2013;27 (2):161-173. doi: 10.1016/j.devcel.2013.09.024. PubMed PMID:24176641 PubMed Central PMC3869996.
- Merle, N, Féraud, O, Gilquin, B, Hubstenberger, A, Kieffer-Jacquinot, S, Assard, N et al.. ATAD3B is a human embryonic stem cell specific mitochondrial protein, re-expressed in cancer cells, that functions as dominant negative for the ubiquitous ATAD3A. Mitochondrion. 2012;12 (4):441-8. doi: 10.1016/j.mito.2012.05.005. PubMed PMID:22664726 .
- Hubstenberger, A, Cameron, C, Shtofman, R, Gutman, S, Evans, TC. A network of PUF proteins and Ras signaling promote mRNA repression and oogenesis in C. elegans. Dev Biol. 2012;366 (2):218-31. doi: 10.1016/j.ydbio.2012.03.019. PubMed PMID:22542599 PubMed Central PMC3361503.
- Gilquin, B, Cannon, BR, Hubstenberger, A, Moulouel, B, Falk, E, Merle, N et al.. The calcium-dependent interaction between S100B and the mitochondrial AAA ATPase ATAD3A and the role of this complex in the cytoplasmic processing of ATAD3A. Mol Cell Biol. 2010;30 (11):2724-36. doi: 10.1128/MCB.01468-09. PubMed PMID:20351179 PubMed Central PMC2876520.
- Hubstenberger, A, Merle, N, Charton, R, Brandolin, G, Rousseau, D. Topological analysis of ATAD3A insertion in purified human mitochondria. J Bioenerg Biomembr. 2010;42 (2):143-50. doi: 10.1007/s10863-010-9269-8. PubMed PMID:20349121 .
- Gilquin, B, Taillebourg, E, Cherradi, N, Hubstenberger, A, Gay, O, Merle, N et al.. The AAA+ ATPase ATAD3A controls mitochondrial dynamics at the interface of the inner and outer membranes. Mol Cell Biol. 2010;30 (8):1984-96. doi: 10.1128/MCB.00007-10. PubMed PMID:20154147 PubMed Central PMC2849464.
- Hubstenberger, A, Labourdette, G, Baudier, J, Rousseau, D. ATAD 3A and ATAD 3B are distal 1p-located genes differentially expressed in human glioma cell lines and present in vitro anti-oncogenic and chemoresistant properties. Exp Cell Res. 2008;314 (15):2870-83. doi: 10.1016/j.yexcr.2008.06.017. PubMed PMID:18639545 .
2017-19 - Young team leader starting grant, ATIP-AVENIR (CNRS-INSERM)
2018 - Award of the Academy of sciences (Académie des sciences, Institut de france), “Les Grandes Avancées Françaises en Biologie”

The 2022 price for the best thesis awarded to Andres Cardona by the Life and Health Sciences doctoral school !
Read More

UCA annual Award Ceremony: 7 iBV members recognised for their scientific contributions
Read More
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
