
Florence BESSE
Post-transcriptional control of neuronal plasticity
Main interests
- Understand how post-transcriptional regulatory processes control neuronal plasticity in vivo
- Study the regulatory mechanisms underlying neuronal RNP granule transport and dynamic properties
- Unravel the functions of mRNA transport and local translation in brain maturation and learning and memory processes
- Combine a variety of complementary approaches and perform multi-scale analyses
- Presentation
- Team Members
- Publications
- Job Offers
- Prizes and Awards
- News
- Team Location
- F. Besse Biography
Scientific Questions
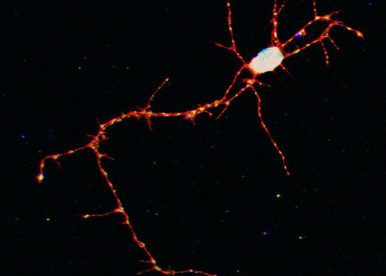
Our main research goal is to understand how post-transcriptional regulatory processes control neuronal plasticity in vivo. Specifically, we study how neuronal ribonucleoprotein (RNP) granules are regulated in space and time to promote local remodeling of neuronal cells during development, as well as in learning and memory processes. Neuronal RNP granules are dynamic macromolecular complexes that contain RNAs and regulatory proteins, assemble in the cell body, and are transported over long-distances to axons or dendrites. They are implicated in the transport of mRNAs, and in their local translation in response to external cues. Although alterations in the properties of neuronal RNP granules have been associated with different neurodegenerative diseases, surprisingly little is known about the molecular mechanisms underlying their formation and dynamics, as well as their physiological function and regulation.
Figure 1 – Neuronal RNP granules
Our Strategy
To understand the role of neuronal RNP granules in neuronal plasticity, as well as their spatio-temporal regulation in response to developmental signals or neuronal activity, one needs to i) dissect the molecular and cellular mechanisms underlying the assembly, transport and remodeling of these granules, and ii) test their functional impact during nervous system development or learning and memory processes. To address these questions, our lab performs multi-scale analyses, and uses Drosophila nervous system as an original in vivo system where advanced genetics, imaging and biochemistry can be combined. We use and develop a range of complementary assays including in vivo live-cell imaging, smFISH, in vivo RIP-seq, high throughput-microscopy, in vitro reconstitution assays. For some of our projects, we collaborate with computer scientists to perform automatic and quantitative image processing, and to develop mathematical models of the biological processes we study (See Morpheme Team ).
Figure 2 – GFP-Imp particle dynamic axonal transport
Research Aims
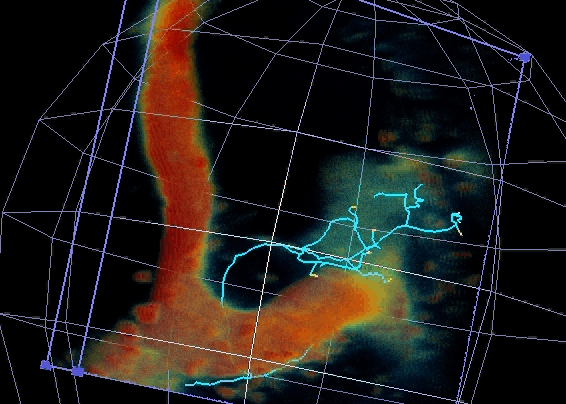
1- Neuronal RNP granules and axonal remodeling. We have shown that RNP granule components are actively recruited to axons undergoing remodeling in response to developmental cues. Furthermore, they are required for the regrowth of axonal processes that occurs after pruning of immature branches. Our objectives are i) to identify the mechanisms triggering RNP granule axonal recruitment and local translation of transported mRNAs, and ii) to understand their impact on axon regrowth and branching.
Figure 3 – 3D reconstruction of a single axon
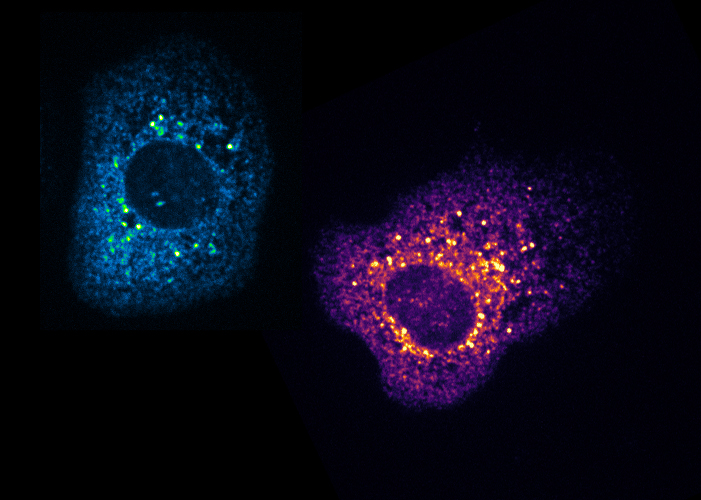
2- Regulation of neuronal RNP granule assembly and dynamics. RNP granules are high order assemblages composed of RNAs and proteins dynamically exchanging with the cytoplasm. To identify regulators of the clustering and recycling of RNP granules, we are combining different approaches including biochemical purifications of RNP complexes, high throughput microscopy-based RNAi screens and in vitro reconstitution assays.
Figure 4 – Granules in cell culture

3- Regulation and function of neuronal RNP granules in synaptic plasticity and disease. We aim at understanding how neuronal RNP granules remodel in response to synaptic activity, and how this contributes to the structural changes underlying the establishment and retention of memories. As the progression of several neurodegenerative diseases has been linked to the formation of pathological RNP aggregates, we also study RNP granules assembled by RNA binding proteins involved in such diseases.
Figure 5 – Drosophila Mushroom Body
Researchers
 DE GRAEVE Fabienne - +33 489150746
DE GRAEVE Fabienne - +33 489150746 MEDIONI Caroline - +33 489150746
MEDIONI Caroline - +33 489150746
PreDocs
 SOLYGA Mathilde - +33 489150746
SOLYGA Mathilde - +33 489150746 NOGUERES Margot - +33 489150746
NOGUERES Margot - +33 489150746 GIRAUD Antoine - +33 489150746
GIRAUD Antoine - +33 489150746
Engineers & Technicians
 PALIN Lucile - +33 489150746
PALIN Lucile - +33 489150746 BLOT Lauren - +33 489150746
BLOT Lauren - +33 489150746 RAMEAU Marion - +33 489150761
RAMEAU Marion - +33 489150761 LE COURTèS Chloé - +33 R
LE COURTèS Chloé - +33 R
Students
 DYSHLEVA Oleksandra - +33 R
DYSHLEVA Oleksandra - +33 R BOUTEILLER PETIT Ilana - +33 R
BOUTEILLER PETIT Ilana - +33 R
Recent publications
- de Queiroz, BR, Laghrissi, H, Rajeev, S, Blot, L, De Graeve, F, Dehecq, M et al.. Axonal RNA localization is essential for long-term memory. Nat Commun. 2025;16 (1):2560. doi: 10.1038/s41467-025-57651-7. PubMed PMID:40089499 PubMed Central PMC11910521.
- Solyga, M, Besse, F. Attenuating the neuronal response to chronic stress through transcription factor aggregation. Trends Neurosci. 2025;48 (4):245-246. doi: 10.1016/j.tins.2025.02.007. PubMed PMID:40069073 .
- Solyga, M, Majumdar, A, Besse, F. Regulating translation in aging: from global to gene-specific mechanisms. EMBO Rep. 2024;25 (12):5265-5276. doi: 10.1038/s44319-024-00315-2. PubMed PMID:39562712 PubMed Central PMC11624266.
- Graeve, F, Debreuve, E, Pushpalatha, KV, Zhang, X, Rahmoun, S, Kozlowski, D et al.. An image-based RNAi screen identifies the EGFR signaling pathway as a regulator of Imp RNP granules. J Cell Sci. 2024;137 (23):. doi: 10.1242/jcs.262119. PubMed PMID:39479884 PubMed Central PMC11698055.
- Garcia-Vaquero, ML, Heim, M, Flix, B, Pereira, M, Palin, L, Marques, TM et al.. Analysis of asymptomatic Drosophila models for ALS and SMA reveals convergent impact on functional protein complexes linked to neuro-muscular degeneration. BMC Genomics. 2023;24 (1):576. doi: 10.1186/s12864-023-09562-4. PubMed PMID:37759179 PubMed Central PMC10523761.
- Bauer, KE, de Queiroz, BR, Kiebler, MA, Besse, F. RNA granules in neuronal plasticity and disease. Trends Neurosci. 2023;46 (7):525-538. doi: 10.1016/j.tins.2023.04.004. PubMed PMID:37202301 .
- Heim, M, Blot, L, Besse, F. An RNA-immunoprecipitation protocol to identify RNAs associated with RNA-binding proteins in cytoplasmic and nuclear Drosophila head fractions. STAR Protoc. 2022;3 (2):101415. doi: 10.1016/j.xpro.2022.101415. PubMed PMID:35634357 PubMed Central PMC9136351.
- Pushpalatha, KV, Solyga, M, Nakamura, A, Besse, F. RNP components condense into repressive RNP granules in the aging brain. Nat Commun. 2022;13 (1):2782. doi: 10.1038/s41467-022-30066-4. PubMed PMID:35589695 PubMed Central PMC9120078.
- Medioni, C, Vijayakumar, J, Ephrussi, A, Besse, F. High-Resolution Live Imaging of Axonal RNP Granules in Drosophila Pupal Brain Explants. Methods Mol Biol. 2022;2431 :451-462. doi: 10.1007/978-1-0716-1990-2_24. PubMed PMID:35412292 .
- De Graeve, F, Formicola, N, Pushpalatha, KV, Nakamura, A, Debreuve, E, Descombes, X et al.. Detecting Stress Granules in Drosophila Neurons. Methods Mol Biol. 2022;2428 :229-242. doi: 10.1007/978-1-0716-1975-9_14. PubMed PMID:35171483 .
- Medioni, C, Ephrussi, A, Besse, F. Live-Imaging of Axonal Cargoes in Drosophila Brain Explants Using Confocal Microscopy. Methods Mol Biol. 2022;2417 :19-28. doi: 10.1007/978-1-0716-1916-2_2. PubMed PMID:35099788 .
- Formicola, N, Heim, M, Dufourt, J, Lancelot, AS, Nakamura, A, Lagha, M et al.. Correction: Tyramine induces dynamic RNP granule remodeling and translation activation in the Drosophila brain. Elife. 2021;10 :. doi: 10.7554/eLife.70755. PubMed PMID:34047275 PubMed Central PMC8163498.
- Formicola, N, Heim, M, Dufourt, J, Lancelot, AS, Nakamura, A, Lagha, M et al.. Tyramine induces dynamic RNP granule remodeling and translation activation in the Drosophila brain. Elife. 2021;10 :. doi: 10.7554/eLife.65742. PubMed PMID:33890854 PubMed Central PMC8064753.
- Pushpalatha, KV, Besse, F. Local Translation in Axons: When Membraneless RNP Granules Meet Membrane-Bound Organelles. Front Mol Biosci. 2019;6 :129. doi: 10.3389/fmolb.2019.00129. PubMed PMID:31824961 PubMed Central PMC6882739.
- Genovese, S, Clément, R, Gaultier, C, Besse, F, Narbonne-Reveau, K, Daian, F et al.. Coopted temporal patterning governs cellular hierarchy, heterogeneity and metabolism in Drosophila neuroblast tumors. Elife. 2019;8 :. doi: 10.7554/eLife.50375. PubMed PMID:31566561 PubMed Central PMC6791719.
- De Graeve, F, Debreuve, E, Rahmoun, S, Ecsedi, S, Bahri, A, Hubstenberger, A et al.. Detecting and quantifying stress granules in tissues of multicellular organisms with the Obj.MPP analysis tool. Traffic. 2019;20 (9):697-711. doi: 10.1111/tra.12678. PubMed PMID:31314165 .
- Formicola, N, Vijayakumar, J, Besse, F. Neuronal ribonucleoprotein granules: Dynamic sensors of localized signals. Traffic. 2019;20 (9):639-649. doi: 10.1111/tra.12672. PubMed PMID:31206920 .
- Vijayakumar, J, Perrois, C, Heim, M, Bousset, L, Alberti, S, Besse, F et al.. The prion-like domain of Drosophila Imp promotes axonal transport of RNP granules in vivo. Nat Commun. 2019;10 (1):2593. doi: 10.1038/s41467-019-10554-w. PubMed PMID:31197139 PubMed Central PMC6565635.
- Razetti, A, Medioni, C, Malandain, G, Besse, F, Descombes, X. A stochastic framework to model axon interactions within growing neuronal populations. PLoS Comput Biol. 2018;14 (12):e1006627. doi: 10.1371/journal.pcbi.1006627. PubMed PMID:30507939 PubMed Central PMC6292646.
- De Graeve, F, Besse, F. Neuronal RNP granules: from physiological to pathological assemblies. Biol Chem. 2018;399 (7):623-635. doi: 10.1515/hsz-2018-0141. PubMed PMID:29641413 .
2024 - Prix Rachel Ajzen et Leon Lagolnitzer de la Fondation pour la Recherche Médicale
2023 - Elected EMBO member
2009 - HFSP Career Development Award
2008 - ATIP Biologie du Développement (CNRS)
2003 - HFSP, EMBO and FEBS Long-term Fellowships

The Localization of RNAs in Neurons: A Key Mechanism for Long-Term Memory
Read More

Florence Besse receives the Rachel Ajzen and Léon Lagolnitzer Prize!
Read More

Florence Besse, élue membre de l’European Molecular Biology Organisation (EMBO)
Read More
Seminar: Travis THOMSON (Wed 1 Feb, 2 pm)
Read More

Tyramine induces dynamic RNP granule remodeling and translation activation in the Drosophila brain
Read More

The 2018 annual PhD prize was awarded to Jeshlee Cyril VIJAYAKUMAR
Read More

When iBV researchers meet Art at the Museum of Modern Art
Read More
From disease to transport in neuronal cells ‘s remodeling, the major role played by a prion-like domain
Read More
iBV - Institut de Biologie Valrose
"Centre de Biochimie"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
Florence Besse received her PhD from Paris University and did her postdoctoral work at EMBL (Heidelberg, Germany) in the lab of Anne Ephrussi, where she started exploring the regulatory mechanisms underlying mRNA localization in the Drosophila nervous system. She established her lab in 2009 at the Institute of Biology Valrose (iBV, Nice, France), funded by competitive ATIP and HFSP career development award starting grants. She is now Research Director at the CNRS and head of the iBV. Florence Besse lab is interested in the spatio-temporal regulation of RNA in the developing, mature and aging Drosophila brain, and in particular in the machineries and physiological signaling pathways that control RNA targeting and translation control. Combining advanced imaging, biochemical purification and transcriptome-wide profiling, her group has recently explored the dynamic regulation and function of RNA condensates in the context of memory circuits.


