
GCE
Genetic Code Expansion
Mission : To support biologists in applying Genetic Code Expansion for advance protein engineering in biological studies.
We provide an easy access to GCE and help with adapting and applying various GCE techniques to fit your needs.
Expertise :
Genetic code expansion (GCE) is one of the most powerful synthetic biology technology for protein engineering. GCE allows site-specific incorporation of non-canonical amino acids (ncAAs) with desired properties into proteins expressed in cells and therefore the ability to manipulate proteins in a precise manner. This is extremely useful for studying protein structure and function in biological processes and for generating proteins and organisms with new properties.
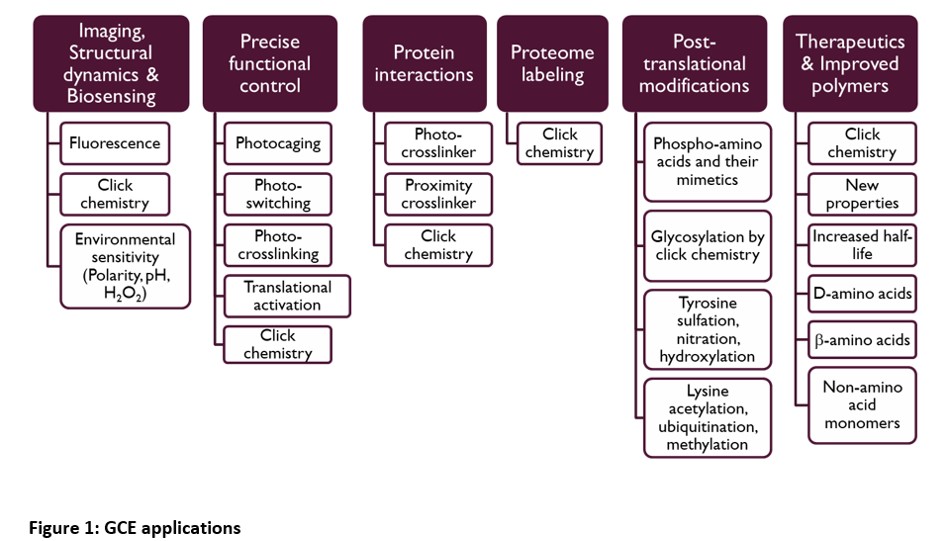
To date, over 200 ncAAs have been incorporated into proteins via GCE. The utilities of GCE span basic science to clinical applications.
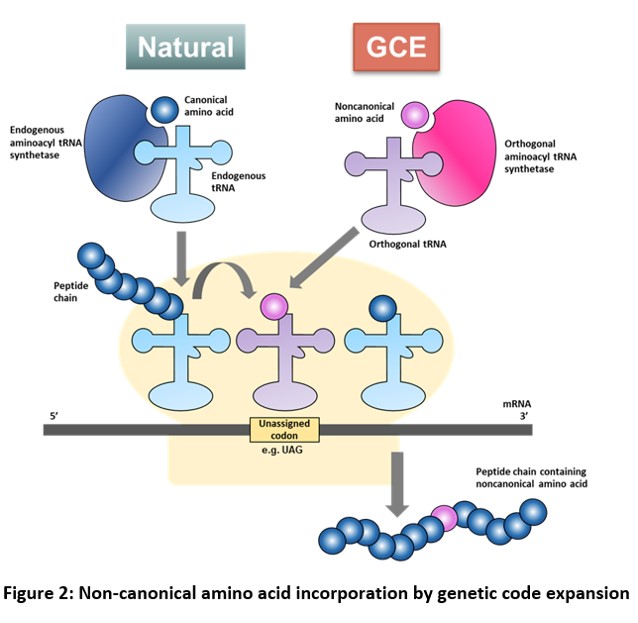
GCE via Stop codon suppression
During normal protein translation, each codon in an mRNA is recognized by a specific anticodon of a tRNA that is aminoacylated with the corresponding amino acid by an aminoacyl tRNA synthetase (aaRS). The ribosome moves along the mRNA and facilitates both the decoding of triplet codons and the polymerization of the corresponding amino acids into a polypeptide. Once a stop codon, which is not recognized by any tRNA, is encountered, the peptide is released. On the other hand, GCE uses an ‘orthogonal’ aaRS-tRNA pair (an aaRS that does not aminoacylate the normal tRNA of a cell and a tRNA that is not a substrate for the normal aaRS of the cell) to direct the incorporation of a ncAA in response to an unassigned codon such as an amber stop codon (TAG) in a gene of interest.
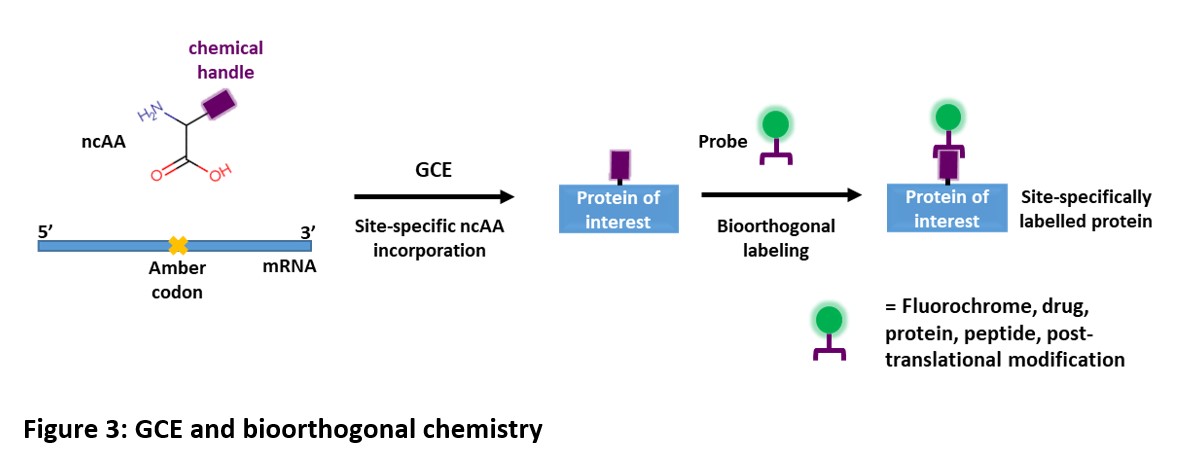
Extended power of GCE via bioorthogonal chemistry
Bioorthogonal chemistry involves chemoselective reactions that can occur inside of living systems without interfering with native biochemical processes. This process is also commonly known as ‘click chemistry’.
Combining the power of GCE and bioorthogonal chemistry can greatly increase the chemistries of the protein. With GCE, it is now easier to add to proteins a range of probes and modifications by cotranslational incorporation of ncAAs bearing functional groups (chemical handles) that can be conjugated with externally added molecules using chemoselective reactions.
Examples of these molecules for bioorthogonal reactions are fluorescent probes, biotin, proteins, lipids, etc. which can be used for fluorescent imaging, purification, creating chimera, adding post-translational modification, etc.
References:
- Chin JW. Expanding and reprogramming the genetic code. Nature. 2017 Oct 4;550(7674):53-60. doi: 10.1038/nature24031. PMID: 28980641.
- Davis L, Chin JW. Designer proteins: applications of genetic code expansion in cell biology. Nat Rev Mol Cell Biol. 2012 Feb 15;13(3):168-82. doi: 10.1038/nrm3286. PMID: 22334143.
- Krall N, da Cruz FP, Boutureira O, Bernardes GJ. Site-selective protein-modification chemistry for basic biology and drug development. Nat Chem. 2016 Feb;8(2):103-13. doi: 10.1038/nchem.2393. Epub 2015 Nov 30. PMID: 26791892.
Platform’s services and collaborations :
The platform provides support starting from GCE project development up to project finalization.
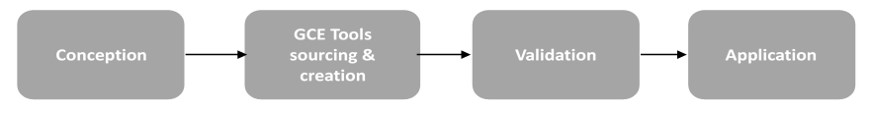
We help you plan the project, supply GCE tools and execute GCE technology. Our activities include:
- Application of existing or customized GCE tools
- Small scale production of ncAA-modified proteins
- Application of bioorthogonal chemistry
Examples of common GCE projects are site-specific incorporation of:
- Photocrosslinking amino acid for studying protein-protein interactions and proteomics analysis
- Clickable amino acid for fluorescent protein labelling
- Clickable amino acid for protein purification
- ncAA for precise translational control of protein expression
Technical manager
Krittalak Chakrabandhu, PhD
Tel : +33 4 89 15 07 94
E-mail : Krittalak.CHAKRABANDHU@univ-cotedazur.fr
Scientific coordinators
Bruno Hudry, PhD
Tel : +33 4 89 15 07 60
E-mail : Bruno.HUDRY@univ-cotedazur.fr
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
