
Thierry LEPAGE
Gene regulatory networks, axis specification and morphogenesis of the sea urchin embryo
Main interests
- Understanding how morphogenesis of the embryo is encoded in the sequence of DNA.
- Understanding how morphogens of the TGF beta family work to pattern the embryo.
- Dissecting out the role of maternal determinants and zygotic genes in axis formation.
- Understanding the logic and the evolution of complex gene regulatory networks driving early development
Scientific Questions
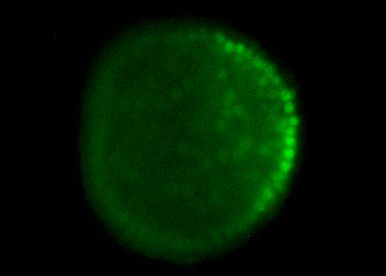
Our goal is to answer fundamental questions such as how an embryo with multiple differentiated cell types can develop from the egg and how is the linear information encoded in the DNA used to shape the embryo during gastrulation and morphogenesis. We try to understand how the main axes of polarity (dorsal-ventral and left-right) are specified and what are the respective roles of the maternal information stored in the egg and of intercellular communication in these processes.
We are also interested in the mechanisms regulating morphogenesis of the larval skeleton, a process that requires epithelial-mesenchymal transition (EMT) and oriented migration of skeletogenic precursors cells located at the vegetal pole of the embryo. We try to understand how beta catenin and MAPK signaling act together with transcription factors to control delamination of these cells and how signals emitted by the ectoderm regulate migration of these mesenchymal cells and control patterning of the skeleton.
Our Strategy
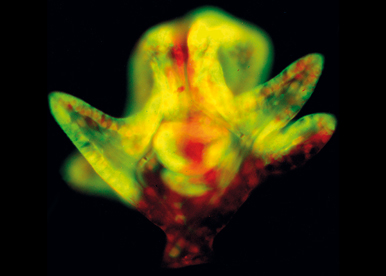
We exploit the power of functional analysis and molecular genetics in the developing sea urchin embryo to dissect the processes of axis formation and morphogenesis. Echinoderms are phylogenetically close to vertebrates and their embryos offer many advantages for the functional analysis of developmental genes. In particular, sea urchin embryos are abundant, fully transparent, develop rapidly and synchronously and are amenable to various experimental perturbations such as mRNA overexpression, microinjection of antisense morpholino oligonucleotides or of CRISPER-Cas9 RNAs, or cell transplantation.
Our strategy is based on the analysis of key signaling molecules and transcription factors. In particular, we analyze the roles of morphogens such as Nodal, BMPs and FGFs that play crucial roles in formation of the dorsal-ventral and left-right axes and of kinases and transcription factors of the homeobox and ETS families that are critical regulators of EMT and morphogenesis of skeleton. We use a combination of biochemistry, molecular genetics and imaging to dissect the function of individual genes in these process as well as large scale approaches and genome wide screens such as RNA-seq to reconstruct the Gene Regulatory Networks that orchestrate these processes.
As a broad approach to sea urchin development, we are also doing a large-scale in situ screen which has already generated a number of useful markers for studying specification of cell fates and patterning of the embryo.
Research Aims
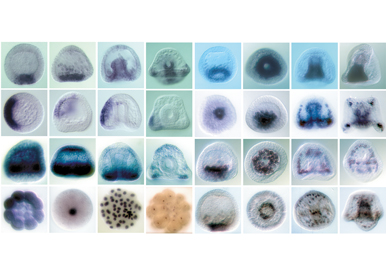
Dissect and model the D/V gene regulatory network activated by Nodal and BMP2/4
One objective is to characterize the maternal factors that shape the Nodal and BMP gradients.To reach this goal, we analyze the function of several maternal factors that we recently identified and that appear to be critical regulators of nodal expression. A second objective is to use RNA-seq screens to extend the gene regulatory network activated by Nodal and BMP2/4 and to construct a logical model of this GRN.
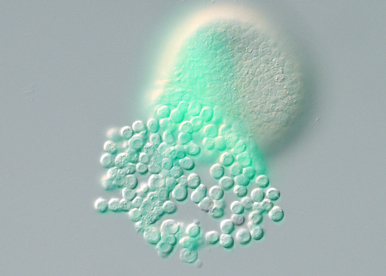
Identify the signals that activate MAP kinase signaling and that regulate EMT and morphogenesis of the skeleton.
One objective is to identify the signals activated by beta catenin signaling that trigger MAPK signaling in the skeletogenic mesoderm. Another objective is to identify the key targets phosphorylated by ERK and the transcription factors that orchestrate the EMT genetic program and morphogenesis of the skeleton.
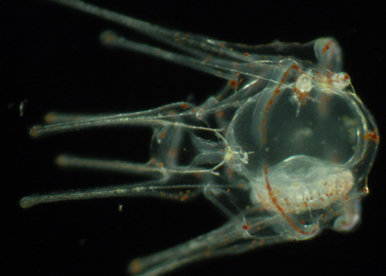
Dissect the early steps leading to establishment of left right asymmetry.
The main objective is to identify the signals that control establishment of the left-right mesendodermal organizer and to understand how the activity of the H+/K+ proton pump, of the Notch/Delta, BMP and FGF signaling pathways act in concert to establish the asymmetrical expression of nodal.
Engineers & Technicians
 CHESSEL Aline - +33 489150831
CHESSEL Aline - +33 489150831
Students
 LOPES Alice - +33 R
LOPES Alice - +33 R CASTEL Océane - +33 A
CASTEL Océane - +33 A
Recent Publications
- Viswanathan, PK, Chessel, A, Molina, MD, Haillot, E, Lepage, T. Maternal TGF-β ligand Panda breaks the radial symmetry of the sea urchin embryo by antagonizing the Nodal type II receptor ACVRII. PLoS Biol. 2024;22 (6):e3002701. doi: 10.1371/journal.pbio.3002701. PubMed PMID:38913712 PubMed Central PMC11239237.
- Chessel, A, De Crozé, N, Molina, MD, Taberner, L, Dru, P, Martin, L et al.. RAS-independent ERK activation by constitutively active KSR3 in non-chordate metazoa. Nat Commun. 2023;14 (1):3970. doi: 10.1038/s41467-023-39606-y. PubMed PMID:37407549 PubMed Central PMC10322840.
- Marlétaz, F, Couloux, A, Poulain, J, Labadie, K, Da Silva, C, Mangenot, S et al.. Analysis of the P. lividus sea urchin genome highlights contrasting trends of genomic and regulatory evolution in deuterostomes. Cell Genom. 2023;3 (4):100295. doi: 10.1016/j.xgen.2023.100295. PubMed PMID:37082140 PubMed Central PMC10112332.
- Floc'hlay, S, Molina, MD, Hernandez, C, Haillot, E, Thomas-Chollier, M, Lepage, T et al.. Deciphering and modelling the TGF-β signalling interplays specifying the dorsal-ventral axis of the sea urchin embryo. Development. 2021;148 (2):. doi: 10.1242/dev.189944. PubMed PMID:33298464 .
- Molina, MD, Lepage, T. Maternal factors regulating symmetry breaking and dorsal-ventral axis formation in the sea urchin embryo. Curr Top Dev Biol. 2020;140 :283-316. doi: 10.1016/bs.ctdb.2019.10.007. PubMed PMID:32591077 .
- Robert, N, Hammami, F, Lhomond, G, Dru, P, Lepage, T, Schubert, M et al.. A wnt2 ortholog in the sea urchin Paracentrotus lividus. Genesis. 2019;57 (11-12):e23331. doi: 10.1002/dvg.23331. PubMed PMID:31479176 .
- Molina, MD, Gache, C, Lepage, T. Expression of exogenous mRNAs to study gene function in echinoderm embryos. Methods Cell Biol. 2019;151 :239-282. doi: 10.1016/bs.mcb.2018.10.011. PubMed PMID:30948011 .
- Molina, MD, Quirin, M, Haillot, E, De Crozé, N, Range, R, Rouel, M et al.. MAPK and GSK3/ß-TRCP-mediated degradation of the maternal Ets domain transcriptional repressor Yan/Tel controls the spatial expression of nodal in the sea urchin embryo. PLoS Genet. 2018;14 (9):e1007621. doi: 10.1371/journal.pgen.1007621. PubMed PMID:30222786 PubMed Central PMC6160229.
- Molina, MD, Quirin, M, Haillot, E, Jimenez, F, Chessel, A, Lepage, T et al.. p38 MAPK as an essential regulator of dorsal-ventral axis specification and skeletogenesis during sea urchin development: a re-evaluation. Development. 2017;144 (12):2270-2281. doi: 10.1242/dev.152330. PubMed PMID:28507001 .
- Karakostis, K, Zanella-Cléon, I, Immel, F, Guichard, N, Dru, P, Lepage, T et al.. A minimal molecular toolkit for mineral deposition? Biochemistry and proteomics of the test matrix of adult specimens of the sea urchin Paracentrotus lividus. J Proteomics. 2016;136 :133-44. doi: 10.1016/j.jprot.2016.01.001. PubMed PMID:26778142 .
- Lapraz, F, Haillot, E, Lepage, T. A deuterostome origin of the Spemann organiser suggested by Nodal and ADMPs functions in Echinoderms. Nat Commun. 2015;6 :8927. doi: 10.1038/ncomms9927. PubMed PMID:26582589 PubMed Central PMC7188468.
- Lapraz, F, Haillot, E, Lepage, T. A deuterostome origin of the Spemann organiser suggested by Nodal and ADMPs functions in Echinoderms. Nat Commun. 2015;6 :8434. doi: 10.1038/ncomms9434. PubMed PMID:26423516 PubMed Central PMC4600745.
- Haillot, E, Molina, MD, Lapraz, F, Lepage, T. The Maternal Maverick/GDF15-like TGF-β Ligand Panda Directs Dorsal-Ventral Axis Formation by Restricting Nodal Expression in the Sea Urchin Embryo. PLoS Biol. 2015;13 (9):e1002247. doi: 10.1371/journal.pbio.1002247. PubMed PMID:26352141 PubMed Central PMC4564238.
- Molina, MD, de Crozé, N, Haillot, E, Lepage, T. Nodal: master and commander of the dorsal-ventral and left-right axes in the sea urchin embryo. Curr Opin Genet Dev. 2013;23 (4):445-53. doi: 10.1016/j.gde.2013.04.010. PubMed PMID:23769944 .
- Bessodes, N, Haillot, E, Duboc, V, Röttinger, E, Lahaye, F, Lepage, T et al.. Reciprocal signaling between the ectoderm and a mesendodermal left-right organizer directs left-right determination in the sea urchin embryo. PLoS Genet. 2012;8 (12):e1003121. doi: 10.1371/journal.pgen.1003121. PubMed PMID:23271979 PubMed Central PMC3521660.
- Range, R, Lepage, T. Maternal Oct1/2 is required for Nodal and Vg1/Univin expression during dorsal-ventral axis specification in the sea urchin embryo. Dev Biol. 2011;357 (2):440-9. doi: 10.1016/j.ydbio.2011.07.005. PubMed PMID:21782809 .
- Croce, J, Range, R, Wu, SY, Miranda, E, Lhomond, G, Peng, JC et al.. Wnt6 activates endoderm in the sea urchin gene regulatory network. Development. 2011;138 (15):3297-306. doi: 10.1242/dev.058792. PubMed PMID:21750039 PubMed Central PMC3133919.
- Saudemont, A, Haillot, E, Mekpoh, F, Bessodes, N, Quirin, M, Lapraz, F et al.. Ancestral regulatory circuits governing ectoderm patterning downstream of Nodal and BMP2/4 revealed by gene regulatory network analysis in an echinoderm. PLoS Genet. 2010;6 (12):e1001259. doi: 10.1371/journal.pgen.1001259. PubMed PMID:21203442 PubMed Central PMC3009687.
- Duboc, V, Lapraz, F, Saudemont, A, Bessodes, N, Mekpoh, F, Haillot, E et al.. Nodal and BMP2/4 pattern the mesoderm and endoderm during development of the sea urchin embryo. Development. 2010;137 (2):223-35. doi: 10.1242/dev.042531. PubMed PMID:20040489 .
- Lapraz, F, Besnardeau, L, Lepage, T. Patterning of the dorsal-ventral axis in echinoderms: insights into the evolution of the BMP-chordin signaling network. PLoS Biol. 2009;7 (11):e1000248. doi: 10.1371/journal.pbio.1000248. PubMed PMID:19956794 PubMed Central PMC2772021.
2009 - Prix Tregouboff in Marine Biology, French Academy of Sciences
2007 - Prize "Coup d'élan pour la recherche", Bettencourt-Schueller Foundation
1995 - Fellowship Human Frontiers, HFSP
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
