
Bruno HUDRY
Cellular sex and physiology
Main interests
- Sex differences in development, metabolism and behaviour
- Molecular pathways mediating sex chromosome effects
- Evolution of sexual dimorphism
- Anatomy and physiology of the reproductive systems
Scientific Questions
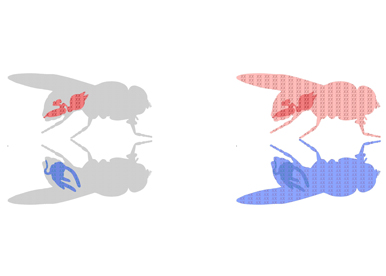
An individual can be characterized by the presence of particular sex organs: testes or ovaries. Formation of these reproductive organs is controlled by specific genetic elements: the sex chromosomes. But sex differences encompass much more than sex organs. Evidence suggests that males and females differ in their normal physiology and susceptibility to a broad range of diseases. These disparities underscore the importance of identifying all the places in the body impacted by sex chromosomes. But besides sex organs we have been operating with a unisex vision of cell and organ functions and implications of sex chromosome presence have been overlooked.
Our long-term objective is to understand where, and how the intrinsic presence of sex chromosomes, cellular sex, drives sex differences in development, physiology, and pathologies. We are currently studying these fundamental questions using Drosophila as an in vivo model.
Our Strategy
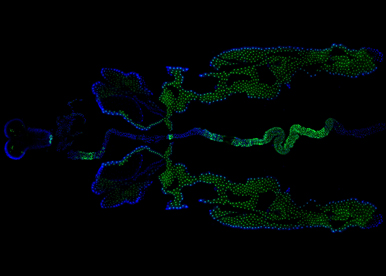
Our group aims to understand how sex chromosome constitution (XX versus XY) impacts physiology across the body. In particular, we want to establish which organs have a sex chromosome set that actively regulates their physiology and how sex chromosome effects are mediated. These questions have been poorly investigated. This is due in part to the difficulty of studying sex chromosome effects in complex higher organisms. In the team, we are using fruit flies to tackle these research questions. Flies have simpler but functionally analogous sex chromosomes and offer the remarkable possibility to generate mosaic animal in which sex chromosomes are genetically manipulated in defined organs. Given the functional similarities between flies and humans, the sex chromosome effects we will uncover in the fly are likely to be relevant to higher organisms and operate in many diseases presenting sexually dimorphic characters.
Our current work aims at identifying new genes/pathways and concepts driving sex chromosome effects using multi-scale approaches combining biochemistry, genetics and cell biology. More specifically, we design genetic and molecular screens to i) identify new target genes downstream of the fly master-switch sex-determining gene, transformer, ii) establish Drosophila as a new paradigm to study somatic loss of the Y chromosome, iii) identify new genes involved in the control of sexual dimorphic trait evolution.
Research Aims
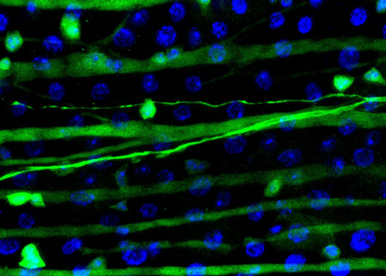
Although our work has demonstrated the impact of the sex determinant transformer in adult intestinal stem cells, the general significance of cellular sex remains poorly understood. In addition, little is known about the molecular mechanisms through which transformer exerts its influence in this new cellular context. Our objectives here are the following: i) to characterise organism-wide where and when cellular sex is required, and ii) to decipher NEW SEX PATHWAYS downstream of transformer.
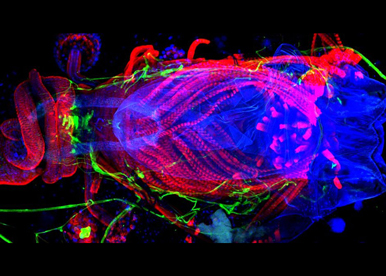
It is widely unknown HOW SEXUAL DIMORPHIC TRAITS CAN EVOLVE so rapidly. In fact, reproductive structures, behaviours and secondary sexual characteristics are some of the most variable and changeable features among insects. These changes may require the rapid evolution rate of the underlying master-switch sex-determining gene. We are currently testing this hypothesis using the sex determinant transformer and closely related Drosophila species as a model
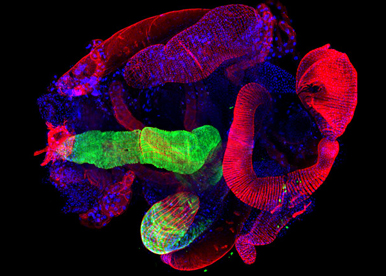
SOMATIC LOSS OF THE Y CHROMOSOME (LOY) is the most common acquired human mutation; its frequency increases with age and is associated with decreased survival time from all causes. Yet the full range of phenotypic consequences of LOY, the genes and mechanisms involved remain elusive. We are developing a technology to achieve for the first time conditional and cell-type specific deletion of entire sex chromosomes. This strategy will be use to model LOY during normal and pathological conditions
Researchers
 DELANOUE Renald - +33 489150763
DELANOUE Renald - +33 489150763 FRANçOIS Charlotte - +33 R
FRANçOIS Charlotte - +33 R TOURNIéRE Océane - +33 489150763
TOURNIéRE Océane - +33 489150763 TOURNIéRE Océane - +33 489150763
TOURNIéRE Océane - +33 489150763 MESCHI Eleonora - +33 R
MESCHI Eleonora - +33 R
Students
 MARIE-SAINTE Eleyna - +33 R
MARIE-SAINTE Eleyna - +33 R
Recent publications
- Hudry, B. Uncovering the mechanisms of sexual differentiation: insights from Drosophila research. C R Biol. 2025;348 :89-105. doi: 10.5802/crbiol.177. PubMed PMID:40464250 .
- Curt, JR, Martín, P, Foronda, D, Hudry, B, Kannan, R, Shetty, S et al.. Ambivalent partnership of the Drosophila posterior class Hox protein Abdominal-B with Extradenticle and Homothorax. PLoS Genet. 2025;21 (1):e1011355. doi: 10.1371/journal.pgen.1011355. PubMed PMID:39804927 PubMed Central PMC11759358.
- Hérault, C, Pihl, T, Hudry, B. Cellular sex throughout the organism underlies somatic sexual differentiation. Nat Commun. 2024;15 (1):6925. doi: 10.1038/s41467-024-51228-6. PubMed PMID:39138201 PubMed Central PMC11322332.
- Clot, C, Hudry, B, Delanoue, R. [The Y chromosome has no impact on sex-specific longevity]. Med Sci (Paris). 2024;40 (2):143-145. doi: 10.1051/medsci/2023203. PubMed PMID:38411420 .
- François, CM, Pihl, T, Dunoyer de Segonzac, M, Hérault, C, Hudry, B. Metabolic regulation of proteome stability via N-terminal acetylation controls male germline stem cell differentiation and reproduction. Nat Commun. 2023;14 (1):6737. doi: 10.1038/s41467-023-42496-9. PubMed PMID:37872135 PubMed Central PMC10593830.
- Delanoue, R, Clot, C, Leray, C, Pihl, T, Hudry, B. Y chromosome toxicity does not contribute to sex-specific differences in longevity. Nat Ecol Evol. 2023;7 (8):1245-1256. doi: 10.1038/s41559-023-02089-7. PubMed PMID:37308701 PubMed Central PMC10406604.
- Banreti, A, Bhattacharya, S, Wien, F, Matsuo, K, Réfrégiers, M, Meinert, C et al.. Biological effects of the loss of homochirality in a multicellular organism. Nat Commun. 2022;13 (1):7059. doi: 10.1038/s41467-022-34516-x. PubMed PMID:36400783 PubMed Central PMC9674851.
- Li, H, Janssens, J, De Waegeneer, M, Kolluru, SS, Davie, K, Gardeux, V et al.. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly. Science. 2022;375 (6584):eabk2432. doi: 10.1126/science.abk2432. PubMed PMID:35239393 PubMed Central PMC8944923.
- Millington, JW, Brownrigg, GP, Chao, C, Sun, Z, Basner-Collins, PJ, Wat, LW et al.. Female-biased upregulation of insulin pathway activity mediates the sex difference in Drosophila body size plasticity. Elife. 2021;10 :. doi: 10.7554/eLife.58341. PubMed PMID:33448263 PubMed Central PMC7864645.
- Duffraisse, M, Paul, R, Carnesecchi, J, Hudry, B, Banreti, A, Reboulet, J et al.. Role of a versatile peptide motif controlling Hox nuclear export and autophagy in the Drosophila fat body. J Cell Sci. 2020;133 (18):. doi: 10.1242/jcs.241943. PubMed PMID:32878938 .
- Hudry, B, de Goeij, E, Mineo, A, Gaspar, P, Hadjieconomou, D, Studd, C et al.. Sex Differences in Intestinal Carbohydrate Metabolism Promote Food Intake and Sperm Maturation. Cell. 2019;178 (4):901-918.e16. doi: 10.1016/j.cell.2019.07.029. PubMed PMID:31398343 PubMed Central PMC6700282.
- Romero-Pozuelo, J, Foronda, D, Martín, P, Hudry, B, Merabet, S, Graba, Y et al.. Cooperation of axial and sex specific information controls Drosophila female genitalia growth by regulating the Decapentaplegic pathway. Dev Biol. 2019;454 (2):145-155. doi: 10.1016/j.ydbio.2019.06.014. PubMed PMID:31251896 .
- Grmai, L, Hudry, B, Miguel-Aliaga, I, Bach, EA. Chinmo prevents transformer alternative splicing to maintain male sex identity. PLoS Genet. 2018;14 (2):e1007203. doi: 10.1371/journal.pgen.1007203. PubMed PMID:29389999 PubMed Central PMC5811060.
- Perea, D, Guiu, J, Hudry, B, Konstantinidou, C, Milona, A, Hadjieconomou, D et al.. Ret receptor tyrosine kinase sustains proliferation and tissue maturation in intestinal epithelia. EMBO J. 2017;36 (20):3029-3045. doi: 10.15252/embj.201696247. PubMed PMID:28899900 PubMed Central PMC5641678.
- Hudry, B, Khadayate, S, Miguel-Aliaga, I. The sexual identity of adult intestinal stem cells controls organ size and plasticity. Nature. 2016;530 (7590):344-8. doi: 10.1038/nature16953. PubMed PMID:26887495 PubMed Central PMC4800002.
- Baëza, M, Viala, S, Heim, M, Dard, A, Hudry, B, Duffraisse, M et al.. Inhibitory activities of short linear motifs underlie Hox interactome specificity in vivo. Elife. 2015;4 :. doi: 10.7554/eLife.06034. PubMed PMID:25869471 PubMed Central PMC4392834.
- Duffraisse, M, Hudry, B, Merabet, S. Bimolecular fluorescence complementation (BiFC) in live Drosophila embryos. Methods Mol Biol. 2014;1196 :307-18. doi: 10.1007/978-1-4939-1242-1_19. PubMed PMID:25151172 .
- Boube, M, Hudry, B, Immarigeon, C, Carrier, Y, Bernat-Fabre, S, Merabet, S et al.. Drosophila melanogaster Hox transcription factors access the RNA polymerase II machinery through direct homeodomain binding to a conserved motif of mediator subunit Med19. PLoS Genet. 2014;10 (5):e1004303. doi: 10.1371/journal.pgen.1004303. PubMed PMID:24786462 PubMed Central PMC4006704.
- Hudry, B, Thomas-Chollier, M, Volovik, Y, Duffraisse, M, Dard, A, Frank, D et al.. Molecular insights into the origin of the Hox-TALE patterning system. Elife. 2014;3 :e01939. doi: 10.7554/eLife.01939. PubMed PMID:24642410 PubMed Central PMC3957477.
- Linneweber, GA, Jacobson, J, Busch, KE, Hudry, B, Christov, CP, Dormann, D et al.. Neuronal control of metabolism through nutrient-dependent modulation of tracheal branching. Cell. 2014;156 (1-2):69-83. doi: 10.1016/j.cell.2013.12.008. PubMed PMID:24439370 PubMed Central PMC3898607.
2024 - EMBO Young Investigator
2024 - CNRS Bronze Medal
2023 - EMBO Young Investigator
2022 - Prix de l’Académie des Sciences - Prix du Dr et de Mme Henri Labbé
2018 - Young team leader starting grant, ATIP-AVENIR (CNRS)

Bruno Hudry awarded an ERC Consolidator Grant 2024!
Read More

Bruno Hudry receives the CNRS Bronze Medal!
Read More

Bruno HUDRY nommé EMBO Young Investigator
Read More

UCA Annual Award Ceremony 2019: 4 iBV members recognized this year !
Read More
Intestinal sex differences in sugar metabolism regulates sperm production
Read More
iBV - Institut de Biologie Valrose
"Centre de Biochimie"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
