
- This event has passed.
Matej KRAJNC
30/10/2023 at 11:30
Event Navigation
From: Jozef Stefan Institute, Ljubljana, SLOVENIA
Will give a seminar entitled:
What do embryos have in common with retail companies and Google?
Complex systems are characterized by their mutlidimensionality and intricate interconnections among their components, which leads to emergent system-scale behavioral patterns that cannot be simply predicted from properties of the individual components. Studying these systems requires systematic quantitative approaches, including the employment of well-organized and properly structured data models to represent their static states as well as their temporal dynamics.
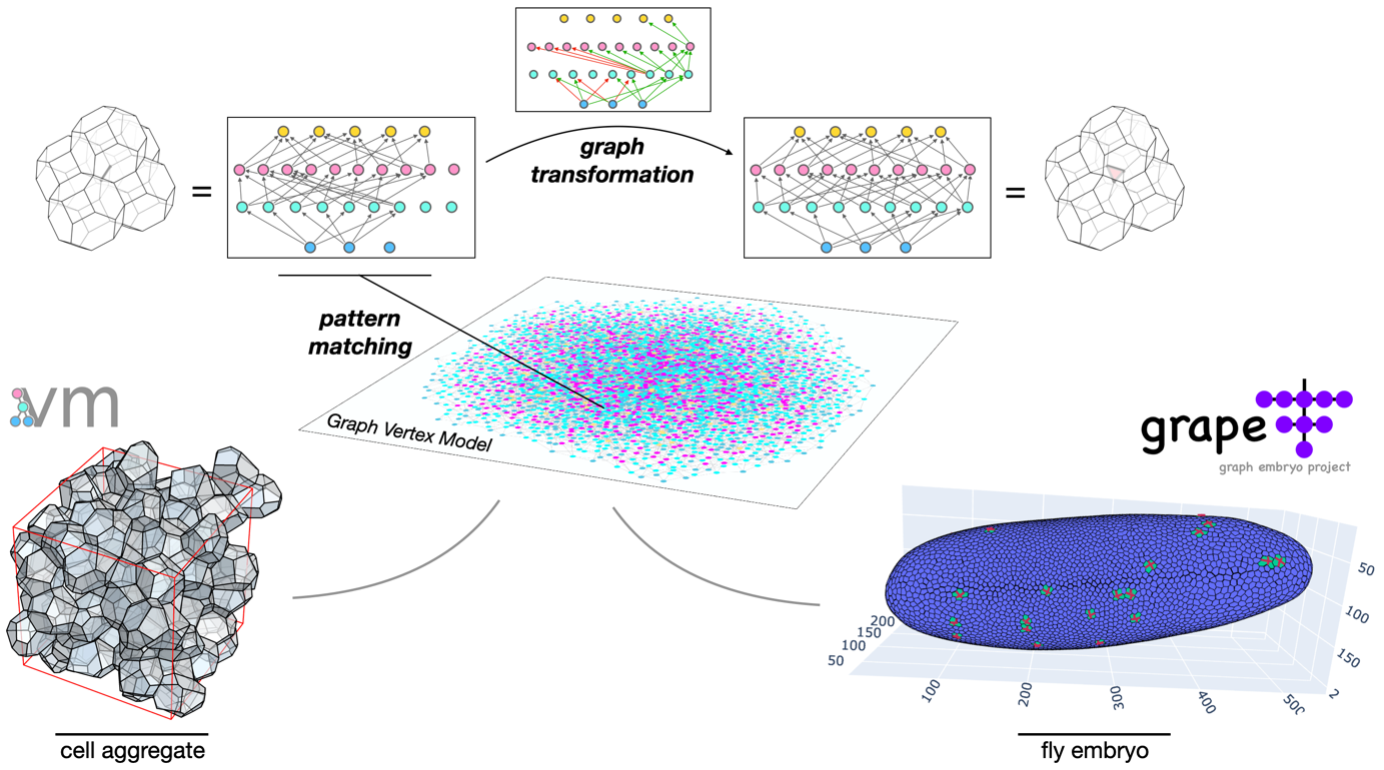
Three-dimensional (3D) cellular aggregates can serve as models of tumors and tumor spheroids and can be computationally studied using the vertex model. It is known that vertex models of 3D cell aggregates with a dynamic topology of the cell network, describing cell reorganizations within the aggregate, are difficult to implement due to the convoluted architecture of the underlying data model. To solve this, we propose a reformulation of the vertex model, called Graph Vertex Model (GVM). GVM stores the topology of the cell network into a knowledge graph, whose data structure, defined by a metagraph, allows capturing the inherent relations between vertices, edges, cell interfaces, and cells. As a result, cell-rearrangement events in GVM simplify to simple graph transformations that are straight-forward to implement and are unified between 2D and 3D space-filling polygonal and polyhedral packings.
We apply a similar approach to live-imaging data of the early fly embryo. To initiate collaborative, interactive, and structured buildup of the common knowledge, we develop GRAPh Embryo project (GRAPE), an online queryable graph database that allows interactive analyses of an entire fly embryo with all surface cells accurately segmented, tracked, and cell divisions identified. Results of queries in GRAPE are automatically projected color-coded onto a 3D image of the embryo, additionally represented in a graph format, and readily downloadable for further analysis. In all, knowledge graphs are potentially a paradigm-shifting concept that offers countless possibilities to develop suitable data models for structured storage, analysis, and manipulation of tissue data.
The MECABIONIC SEMINARS is part of the action of the new MECABIONIC program at UniCA (https://univ-cotedazur.eu/events/mecabionic). The aim of this seminar series is to highlight recent advances and breakthroughs in mechanobiology and to gather researchers tackling the mechanics of the living. We believe this to be a unique opportunity to foster exciting and insightful discussions among a multi- and inter-disciplinary community of experts focused on the understanding of the fundamental biophysical principles powering life from the molecule to the organism level.
In the last decades, biology has been increasingly becoming a science at the cross-road of mathematics, physics, chemistry, engineering and computer science. Therefore, it is key to promote synergistic interaction between scientists working in different disciplines that often have difficulties cross-communicating. With the MECABIONIC SEMINAR series, we intend to break the communication barriers by imposing a pedagogic seminar format adapted to a multi- and inter-disciplinary community of junior and senior scientists.
This seminar event will foster interaction among scientists of different UniCA institutions (iBV, LJAD, IRCAN, IPMC, INPHYNI) and promote new interdisciplinary collaborations.
