
Valentina CIGLIOLA
Innate Mechanisms of Spinal Cord Regeneration
Main interests
- Progenitor Cell Responses to Spinal Cord Injury
- Axon Regeneration and Neurogenesis
- Restoration of Functional Circuits and Motor Function after Spinal Cord Injury
- Cross-Species Variation in Spinal Cord Regenerative Capacity
Scientific Questions
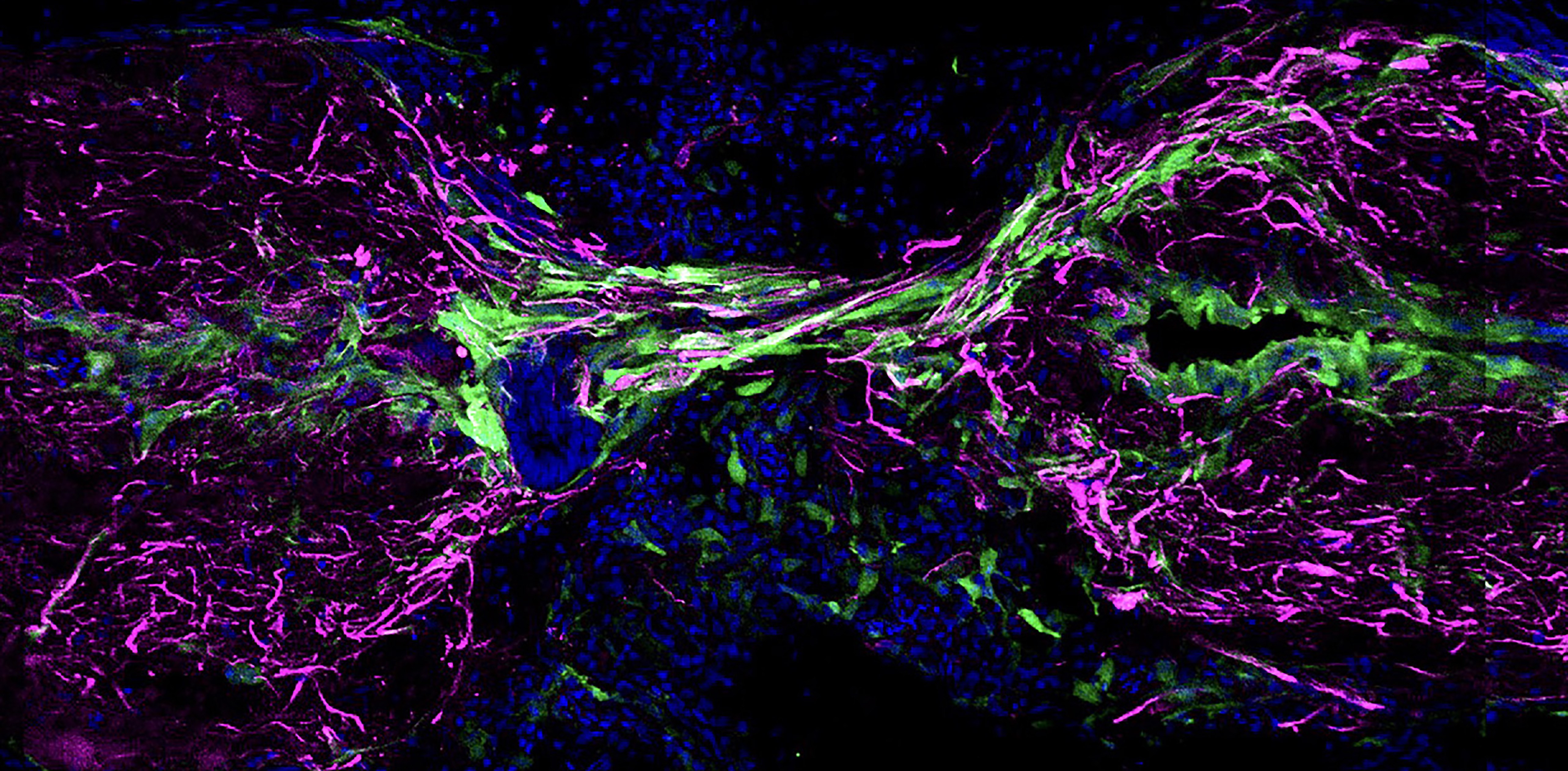
Our main research goal is to understand mechanisms of spinal cord regeneration. While adult mammals (including humans) undergo scarring after spinal cord injury, zebrafish vigorously regenerate and completely recover function after a paralyzing injury. Key for this regenerative capacity are both neurogenesis and the formation of a tissue bridge between the two severed spinal cord stumps, composed of glial cells and nerve fibers (axons). Neonatal mice also regenerate axons after spinal cord injury, and recent studies have bolstered the idea of similar regeneration mechanisms to those in zebrafish. We ask the following questions: What are the factors and mechanisms allowing innate spinal cord regeneration to occur? How are these mechanisms regulated? Are there conserved pro-regenerative mechanisms between zebrafish and neonatal mouse spinal cord? Can regeneration in adult mammals be awaken by re-establishment of innate pro-regenerative mechanisms?
Figure: longitudinal section of zebrafish spinal cord at 2 weeks post transection injury showing tissue bridging between the two severed spinal cord stumps. Glia labeled in magenta and cell nuclei stained in blue.
Our Strategy
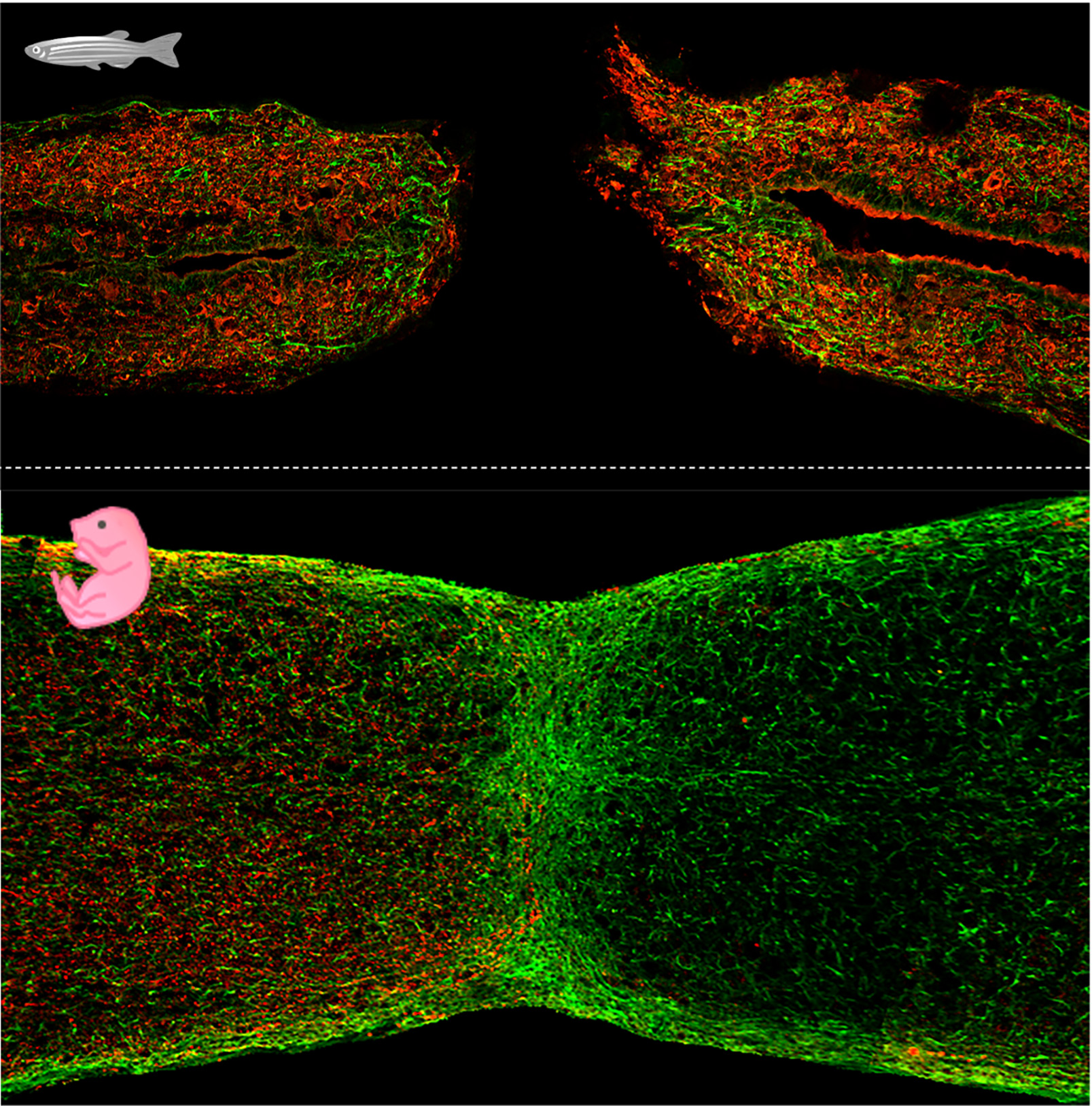
Our primary model system is the zebrafish, possessing many experimental advantages (i.e. large clutches, amenability to mutagenesis) and allowing to apply molecular genetics to spectacular regenerative events. A subset of our studies employs neonatal mice, recently emerged as an informative mammalian model to study natural spinal cord regeneration.
In zebrafish, we combine transcriptomic screens, genetic gain-and-loss of function, regeneration and functional assays to identify factors and mechanisms allowing pro-regenerative responses of progenitor, glial and neuronal cells.
In neonatal mice, we have implemented viral vector approaches to target cells at the site of injury. We combine these approaches with cell ablation and lineage tracing strategies to investigate pro-regenerative responses, and their similarities to those in zebrafish. Ultimately, our aim is to combine these discoveries and develop strategies to awaken regeneration in adult mammals.
Figure: longitudinal sections of zebrafish (top) and neonatal mouse (bottom) spinal cord at 3 days post transection or crush injury, respectively. Glial cells labeled in green, axons labeled in red.
Research Aims
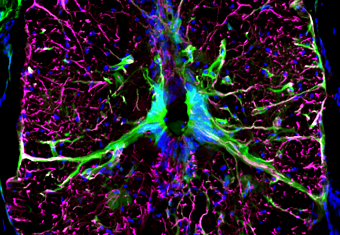
Characterizing the Molecular Mechanisms of Bridge-Associated Neurogenesis
In a recent study, I identified a secreted factor required to activate neurogenesis and axon growth during zebrafish spinal cord regeneration. We are exploring its cellular and molecular mechanisms of action, by developing new methods for imaging of key subcellular, cellular and molecular events during regeneration.
Figure: cross-section of zebrafish spinal cord showing neuron progenitors around the central canal in green. Glia labeled in magenta and cell nuclei stained in blue.
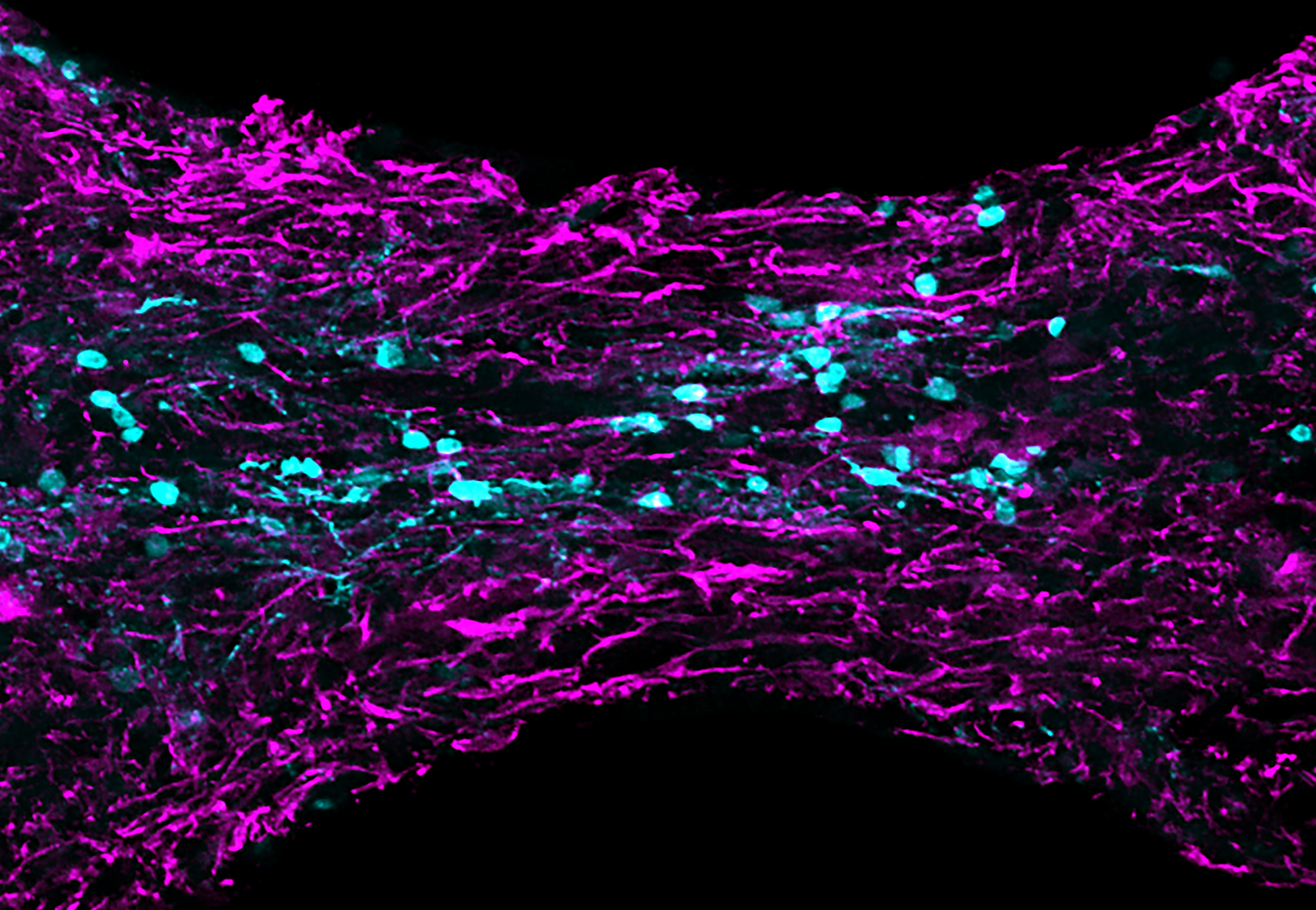
Uncovering New Factors Allowing Zebrafish Spinal Cord Regeneration
We are using transcriptomics, strategies to visualize pro-regenerative cell identities in situ and regeneration assays to pinpoint signals required for zebrafish spinal cord regeneration. We focus on identification of factors guiding progenitor cell responses and growth of axons across the lesion, while keeping an eye open on how to use them in platforms to enhance regenerative capacity in adult mammals.
Figure: longitudinal section of zebrafish spinal cord showing newly generated bridge neurons (cyan) at 3 weeks post injury. Glia labeled in magenta.
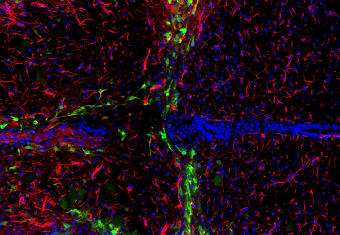
Elucidating Evolutionarily Conserved Spinal Cord Regeneration Responses
In previous studies, I identified zebrafish injury-responsive chromatin regions (regeneration enhancers) functionally conserved in mouse. Notably, for some of them recognition and function are conserved in neonatal mice, regenerating axons as the zebrafish, but not in adult mice undergoing scarring. We are using this knowledge to identify conserved mechanisms of spinal cord regeneration across species, whose re-establishment in adult mammals would possibly boost regeneration.
Figure: neonatal mouse spinal cord at 4 days post crush injury, showing expression driven by zebrafish regeneration enhancers in green. Glia labeled in red and cell nuclei stained in blue.
Postdocs
 LIMA FERNANDES Vania - +33 489150878
LIMA FERNANDES Vania - +33 489150878
Engineers & Technicians
 UVEIRA Julie - +33 489150878
UVEIRA Julie - +33 489150878
Masters
 GIRAUD Antoine - +33 R
GIRAUD Antoine - +33 R
Recent publications
- Becker, CJ, Cigliola, V, Gillotay, P, Rich, A, De Simone, A, Han, Y et al.. In toto imaging of glial JNK signaling during larval zebrafish spinal cord regeneration. Development. 2023;150 (24):. doi: 10.1242/dev.202076. PubMed PMID:37997694 PubMed Central PMC10753585.
- Cigliola, V, Shoffner, A, Lee, N, Ou, J, Gonzalez, TJ, Hoque, J et al.. Spinal cord repair is modulated by the neurogenic factor Hb-egf under direction of a regeneration-associated enhancer. Nat Commun. 2023;14 (1):4857. doi: 10.1038/s41467-023-40486-5. PubMed PMID:37567873 PubMed Central PMC10421883.
- Yan, R, Cigliola, V, Oonk, KA, Petrover, Z, DeLuca, S, Wolfson, DW et al.. An enhancer-based gene-therapy strategy for spatiotemporal control of cargoes during tissue repair. Cell Stem Cell. 2023;30 (1):96-111.e6. doi: 10.1016/j.stem.2022.11.012. PubMed PMID:36516837 PubMed Central PMC9830588.
- Perez-Frances, M, van Gurp, L, Abate, MV, Cigliola, V, Furuyama, K, Bru-Tari, E et al.. Author Correction: Pancreatic Ppy-expressing γ-cells display mixed phenotypic traits and the adaptive plasticity to engage insulin production. Nat Commun. 2021;12 (1):5783. doi: 10.1038/s41467-021-26062-9. PubMed PMID:34580307 PubMed Central PMC8476505.
- Gemberling, MP, Siklenka, K, Rodriguez, E, Tonn-Eisinger, KR, Barrera, A, Liu, F et al.. Transgenic mice for in vivo epigenome editing with CRISPR-based systems. Nat Methods. 2021;18 (8):965-974. doi: 10.1038/s41592-021-01207-2. PubMed PMID:34341582 PubMed Central PMC8349887.
- Oropeza, D, Cigliola, V, Romero, A, Chera, S, Rodríguez-Seguí, SA, Herrera, PL et al.. Stage-specific transcriptomic changes in pancreatic α-cells after massive β-cell loss. BMC Genomics. 2021;22 (1):585. doi: 10.1186/s12864-021-07812-x. PubMed PMID:34340653 PubMed Central PMC8330016.
- Perez-Frances, M, van Gurp, L, Abate, MV, Cigliola, V, Furuyama, K, Bru-Tari, E et al.. Pancreatic Ppy-expressing γ-cells display mixed phenotypic traits and the adaptive plasticity to engage insulin production. Nat Commun. 2021;12 (1):4458. doi: 10.1038/s41467-021-24788-0. PubMed PMID:34294685 PubMed Central PMC8298494.
- Shoffner, A, Cigliola, V, Lee, N, Ou, J, Poss, KD. Tp53 Suppression Promotes Cardiomyocyte Proliferation during Zebrafish Heart Regeneration. Cell Rep. 2020;32 (9):108089. doi: 10.1016/j.celrep.2020.108089. PubMed PMID:32877671 PubMed Central PMC7494019.
- Thompson, JD, Ou, J, Lee, N, Shin, K, Cigliola, V, Song, L et al.. Identification and requirements of enhancers that direct gene expression during zebrafish fin regeneration. Development. 2020;147 (14):. doi: 10.1242/dev.191262. PubMed PMID:32665240 PubMed Central PMC7406312.
- Cigliola, V, Becker, CJ, Poss, KD. Building bridges, not walls: spinal cord regeneration in zebrafish. Dis Model Mech. 2020;13 (5):. doi: 10.1242/dmm.044131. PubMed PMID:32461216 PubMed Central PMC7272344.
- Cigliola, V, Ghila, L, Chera, S, Herrera, PL. Tissue repair brakes: A common paradigm in the biology of regeneration. Stem Cells. 2020;38 (3):330-339. doi: 10.1002/stem.3118. PubMed PMID:31722129 .
- Han, Y, Chen, A, Umansky, KB, Oonk, KA, Choi, WY, Dickson, AL et al.. Vitamin D Stimulates Cardiomyocyte Proliferation and Controls Organ Size and Regeneration in Zebrafish. Dev Cell. 2019;48 (6):853-863.e5. doi: 10.1016/j.devcel.2019.01.001. PubMed PMID:30713073 PubMed Central PMC6435404.
- Cigliola, V, Ghila, L, Thorel, F, van Gurp, L, Baronnier, D, Oropeza, D et al.. Pancreatic islet-autonomous insulin and smoothened-mediated signalling modulate identity changes of glucagon+ α-cells. Nat Cell Biol. 2018;20 (11):1267-1277. doi: 10.1038/s41556-018-0216-y. PubMed PMID:30361701 PubMed Central PMC6215453.
- Berchtold, LA, Miani, M, Diep, TA, Madsen, AN, Cigliola, V, Colli, M et al.. Pannexin-2-deficiency sensitizes pancreatic β-cells to cytokine-induced apoptosis in vitro and impairs glucose tolerance in vivo. Mol Cell Endocrinol. 2017;448 :108-121. doi: 10.1016/j.mce.2017.04.001. PubMed PMID:28390953 .
- Cigliola, V, Populaire, C, Pierri, CL, Deutsch, S, Haefliger, JA, Fadista, J et al.. A Variant of GJD2, Encoding for Connexin 36, Alters the Function of Insulin Producing β-Cells. PLoS One. 2016;11 (3):e0150880. doi: 10.1371/journal.pone.0150880. PubMed PMID:26959991 PubMed Central PMC4784816.
- Cigliola, V, Allagnat, F, Berchtold, LA, Lamprianou, S, Haefliger, JA, Meda, P et al.. Role of Connexins and Pannexins in the Pancreas. Pancreas. 2015;44 (8):1234-44. doi: 10.1097/MPA.0000000000000378. PubMed PMID:26465951 .
- Chera, S, Baronnier, D, Ghila, L, Cigliola, V, Jensen, JN, Gu, G et al.. Diabetes recovery by age-dependent conversion of pancreatic δ-cells into insulin producers. Nature. 2014;514 (7523):503-7. doi: 10.1038/nature13633. PubMed PMID:25141178 PubMed Central PMC4209186.
- Cigliola, V, Chellakudam, V, Arabieter, W, Meda, P. Connexins and β-cell functions. Diabetes Res Clin Pract. 2013;99 (3):250-9. doi: 10.1016/j.diabres.2012.10.016. PubMed PMID:23176806 .
- Nlend, RN, Aït-Lounis, A, Allagnat, F, Cigliola, V, Charollais, A, Reith, W et al.. Cx36 is a target of Beta2/NeuroD1, which associates with prenatal differentiation of insulin-producing β cells. J Membr Biol. 2012;245 (5-6):263-73. doi: 10.1007/s00232-012-9447-1. PubMed PMID:22729650 .
- Potolicchio, I, Cigliola, V, Velazquez-Garcia, S, Klee, P, Valjevac, A, Kapic, D et al.. Connexin-dependent signaling in neuro-hormonal systems. Biochim Biophys Acta. 2012;1818 (8):1919-36. doi: 10.1016/j.bbamem.2011.09.022. PubMed PMID:22001400 .

The Cigliola laboratory invites applications for a Research Technician position at the Institute of Biology Valrose in Nice, France. Our lab uses zebrafish and mouse models to study Innate Mechanisms of Spinal Cord Regeneration. We are looking for highly motivated and ambitious candidates with interest for regenerative biology and neuroscience. Enthusiasm for working with animals is essential. Specialized training will be provided. The candidate will have the opportunity to figure as co-author on published manuscripts.
The Institute of Biology Valrose is a research Center of Excellence at the University of Côte d’Azur and offers a vibrant research environment with national and international scientists and collaborations (iBV). Nice is located on the French Riviera and offers amazing opportunities for outdoor recreation.
Technical duties include:
- Zebrafish breeding, husbandry, and surgeries; molecular biology; generation of mutant or transgenic animals by microinjection.
- Contribution to research studies, experiments, data collection and assays.
- Maintenance of general lab supplies; ordering and preparation of reagents and solutions; assistancewith maintaining equipment and lab reagent inventories.
- Review of laboratory methods manuals.
- Implementation and supervision of laboratory safety procedures.Required qualifications:Bachelor’s degree in biology or a related field and at least 2-years previous experience in a research lab. The qualified candidate should have strong troubleshooting skills, should be motivated to learn new techniques, organized, and able to work effectively in a team environment. Previous experience with zebrafish is preferred (but not required). Candidates should be fluent in English, as it is the working language in the laboratory.
Please send your application in a single PDF, including (1) a cover letter that includes a description of research interests and experience, (2) CV and (3) contact information of 2 or more references to Valentina CIGLIOLA. Review of applications will begin immediately, and the position will remain open until filled.

The Cigliola laboratory invites applications for PhD Student and Postdoc positions at the Institute of Biology Valrose in Nice, France. Our lab uses zebrafish and mouse models to study Mechanisms of Spinal Cord Regeneration.
While in adult mammals (including humans) spinal cord injury causes lifelong paralysis, zebrafish possess an astounding capacity to regenerate and recover function after a paralyzing injury. We are interested in the following questions: What are the factors and mechanisms allowing innate spinal cord regeneration to occur? How are these mechanisms regulated? Can regeneration in adult mammals be enhanced by re-establishment of innate pro-regenerative programs?
We are looking for curiosity-driven, motivated and ambitious candidates with interest for regenerative biology and neuroscience. We strongly value team spirit, a positive work environment and collaborations. Enthusiasm for working with zebrafish and/or mice is essential. Candidates should be fluent in English, as it is the working language in the laboratory.
The Institute of Biology Valrose is a research Center of Excellence at the University of Côte d’Azur and offers a vibrant research environment with national and international scientists and collaborations (iBV). Nice is located on the French Riviera and offers amazing opportunities for outdoor recreation.
Please send your application in a single PDF, including (1) a cover letter that includes a description of research interests and experience, (2) CV and (3) contact information of 2 or more references to Valentina CIGLIOLA. Review of applications will begin immediately, and the position will remain open until filled.
2022 - Labex SIGNALIFE Chairs of Excellence
2022 - Chaire de Professeur Junior Starting Grant, ANR/INSERM
2022 - Scholarship to attend the 3rd Nordic Meeting on Development, Stem Cells and Regeneration, Denmark
2022 - Gordon Conference CNS Injury and Repair, Oxnard, CA, (USA) Poster Prize
2020 - Swiss National Science Foundation Postdoc Mobility Fellowships
2017 - Swiss National Science Foundation Early Postdoc Mobility Fellowship
2017 - Regeneration Next Postdoctoral Fellowship
2017 - Dpt. of Cell Biology, Duke University, Durham, NC USA Best Microscopy Image
2017 - PhD School University of Geneva, Switzerland, Best Thesis Award 2016
2015 - Scholarship to attend the The Islet Study Group & Beta Cell Workshop, Israel
2013 - G2L2 Meeting, Geneva, Switzerland, Poster Prize
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2

