
Michèle STUDER
Physiological and pathological mechanisms of neural development
Main interests
- COUP-TFI/NR2F1 in neurodevelopmental cortical disorders
- Transcriptional regulation and activity-dependent mechanisms during mammalian circuit formation
- Mechanisms controlling topographic neuronal connectivity
- Mapping neuronal populations involved in sensorimotor auditory circuits
Scientific Questions
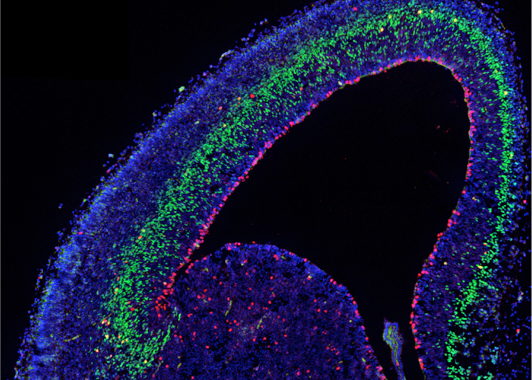
During neurogenesis, different progenitor and neuronal cell types are sequentially generated from a complex population of multipotent stem cells following a precise spatial and temporal pattern before being assembled into maps and circuits. We aim to dissect the cellular and molecular mechanisms by which these different cell types are regulated by defined regionalized and coordinated intrinsic programs and by extrinsic activity-dependent cues that continuously interact during pre- and postnatal development, and control neuronal traits and topographic map formation.
We work on two major regions of the mouse brain: the forebrain and the hindbrain. In the forebrain, we focus on the transcriptional regulator COUP-TFI/NR2F1 recently identified as a neurodevelopmental disease gene and playing multiple roles during neocortical and hippocampal development. In the hindbrain, we aim to identify the different subpopulations arising from rhombomere 4 and contributing to the assembly of the central auditory, trigeminal and vestibular systems.
Fig. 1 – Embryonic mouse cortex labeled with cell-type specific antibodie
Our Strategy
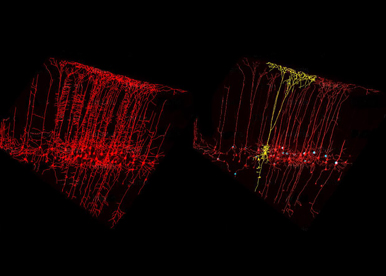
To unravel some of the key molecular and cellular mechanisms underlying neural organization and circuit formation, we use the mouse as an in vivo experimental model and several genetic mutants, which reproduce the clinical features of patients with either intellectual disabilities or hearing sensory impairments. Our strategy is to study the specification of progenitor and neuronal cell types as well as the assembly of neuronal circuits required in shaping distinct functional topographic maps in the healthy and diseased brain.
The team has added to the standard molecular and cellular approaches a whole series of interdisciplinary experimental techniques. We combine genetic gain-and-loss of function approaches, in utero electroporation, 3D imaging of axonal tracts, lineage tracing, in-depth morphological analysis, mouse behavior and high-throughput molecular screening to elucidate how cell specification, migration and connectivity are functionally coordinated and ensure proper assembly of subcircuits in the developing brain. We also aim to develop cerebral 3D organoids as an in vitro model of human development and disease with the overall goal to assess mutations identified in patients.
As developmental abnormalities participate in the etiology of several neuropsychiatric disorders, understanding how different brain populations become organized and connected is essential to advance our comprehension of cognitive human diseases.
Research Aims
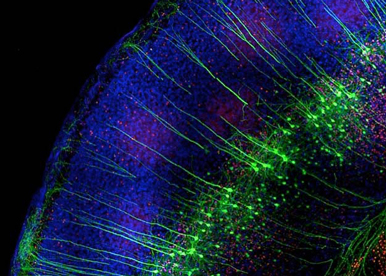
Neurodevelopmental diseases arise from anomalies occurring during the embryonic or fetal age and impairing normal brain functioning. Haploinsufficiency of the orphan nuclear receptor COUP-TFI/NR2F1 leads to developmental delay, intellectual disability, autistic behavior, infantile epilepsy and optic atrophy. We are using different mouse models, in utero electroporation and in vitro approaches to understand the contribution of single mutations identified in patients in the pathophysiological mechanisms of this newly genetic rare disease.
Fig. 3 – Electroporated mouse brain
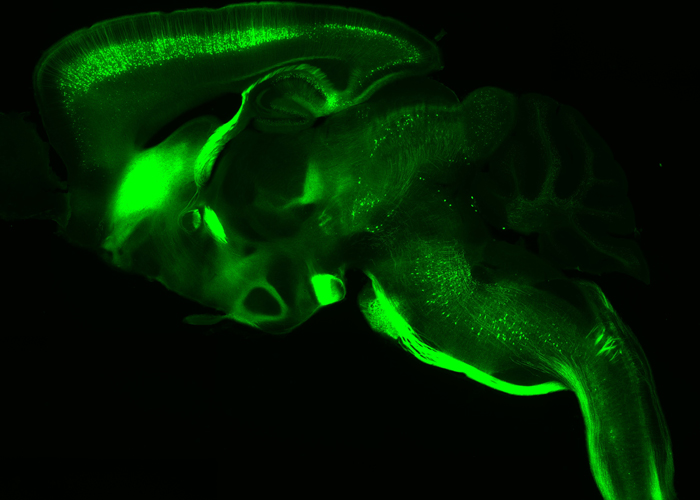
Transcriptional regulation and activity-dependent mechanisms are essential in the maturation of neurons and formation of complex neuronal networks. We are interested in understanding their contribution in the topographic connectivity between the cortex and their subcerebral targets. We focus on the molecular cascade required in driving layer V projection neurons into corticopontine or corticospinal motor neurons, and in remodeling layer IV neurons during the formation of primary somatosensory maps.
Fig. 4 – Thy1-eYFP-H transgenic line
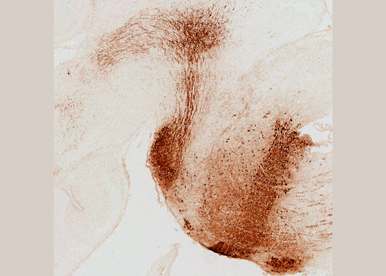
Through functional and intersectional mouse genetics, we aim to map auditory pathways and networks in normal and pathological hearing conditions as well as to identify novel genes involved in the specification of auditory subtypes in the central nervous system. In particular, we will investigate how cells originating from distinct subdomains of rhombomere 4 (r4) contribute in shaping coordinated functional sensorimotor auditory subcircuits.
Fig. 5 - Rhombomere 4 fate map
Researchers
 CHASSOT Anne-Amandine - +33 489150726
CHASSOT Anne-Amandine - +33 489150726 D`AUTRéAUX Fabien - +33 489150846
D`AUTRéAUX Fabien - +33 489150846 DESCHAUX Olivier - +33 489150721
DESCHAUX Olivier - +33 489150721 BERTACCHI Michele - +33 489150721
BERTACCHI Michele - +33 489150721
PreDocs
 CASSINI Paul - +33 489150721
CASSINI Paul - +33 489150721  PIOVANI Paolo - +33 489150721
PIOVANI Paolo - +33 489150721 PIOVANI Paolo - +33 489150721
PIOVANI Paolo - +33 489150721
Engineers & Technicians
 MANTILLERI Annabelle - +33 489150723
MANTILLERI Annabelle - +33 489150723 DALLORTO Eleonora - +33 489150721
DALLORTO Eleonora - +33 489150721
Students
 BOLOGNESE Gaia - +33 R
BOLOGNESE Gaia - +33 R LANTERI Léa - +33 R
LANTERI Léa - +33 R LEKCHIRI Islem - +33 R
LEKCHIRI Islem - +33 R
- Maass, JG, Kamionek, D, Mantilleri, A, Theiss, S, Dötsch, L, Franke, F et al.. Models of Bosch-Boonstra-Schaaf optic atrophy syndrome reveal genotype-phenotype correlations in brain structure and behavior. Dis Model Mech. 2025;18 (10):. doi: 10.1242/dmm.052426. PubMed PMID:40855817 PubMed Central PMC12505268.
- Serra, L, Nordin, A, Jonasson, M, Marenco, C, Rovelli, G, Diebels, A et al.. SOX2 and NR2F1 coordinate the gene expression program of the early postnatal visual thalamus. Biol Open. 2025;14 (8):. doi: 10.1242/bio.062014. PubMed PMID:40631402 PubMed Central PMC12352279.
- Bertacchi, M, Maharaux, G, Studer, M. A Hybrid 2D/3D Approach for Neural Differentiation Into Telencephalic Organoids and Efficient Modulation of FGF8 Signaling. Bio Protoc. 2025;15 (12):e5354. doi: 10.21769/BioProtoc.5354. PubMed PMID:40620811 PubMed Central PMC12222629.
- Bertacchi, M, Theiß, S, Ahmed, A, Eibl, M, Loubat, A, Maharaux, G et al.. Unravelling the conundrum of nucleolar NR2F1 localization using antibody-based approaches in vitro and in vivo. Commun Biol. 2025;8 (1):594. doi: 10.1038/s42003-025-07985-1. PubMed PMID:40204944 PubMed Central PMC11982218.
- Øvsthus, M, van Swieten, MMH, Puchades, MA, Tocco, C, Studer, M, Bjaalie, JG et al.. Spatially integrated cortico-subcortical tracing data for analyses of rodent brain topographical organization. Sci Data. 2024;11 (1):1214. doi: 10.1038/s41597-024-04060-y. PubMed PMID:39532918 PubMed Central PMC11557934.
- Bertacchi, M, Maharaux, G, Loubat, A, Jung, M, Studer, M. FGF8-mediated gene regulation affects regional identity in human cerebral organoids. Elife. 2024;13 :. doi: 10.7554/eLife.98096. PubMed PMID:39485283 PubMed Central PMC11581432.
- Bonzano, S, Dallorto, E, Bovetti, S, Studer, M, De Marchis, S. Mitochondrial regulation of adult hippocampal neurogenesis: Insights into neurological function and neurodevelopmental disorders. Neurobiol Dis. 2024;199 :106604. doi: 10.1016/j.nbd.2024.106604. PubMed PMID:39002810 .
- Cui, K, Xia, Y, Patnaik, A, Salivara, A, Lowenstein, ED, Isik, EG et al.. Genetic identification of medullary neurons underlying congenital hypoventilation. Sci Adv. 2024;10 (25):eadj0720. doi: 10.1126/sciadv.adj0720. PubMed PMID:38896627 PubMed Central PMC11186509.
- Marino, V, Phromkrasae, W, Bertacchi, M, Cassini, P, Chakrabandhu, K, Dell'Orco, D et al.. Disrupted protein interaction dynamics in a genetic neurodevelopmental disorder revealed by structural bioinformatics and genetic code expansion. Protein Sci. 2024;33 (4):e4953. doi: 10.1002/pro.4953. PubMed PMID:38511490 PubMed Central PMC10955615.
- Deloulme, JC, Leclercq, M, Deschaux, O, Flore, G, Capellano, L, Tocco, C et al.. Structural interhemispheric connectivity defects in mouse models of BBSOAS: Insights from high spatial resolution 3D white matter tractography. Neurobiol Dis. 2024;193 :106455. doi: 10.1016/j.nbd.2024.106455. PubMed PMID:38408685 .
- Di Bonito, M, Bourien, J, Tizzano, M, Harrus, AG, Puel, JL, Avallone, B et al.. Abnormal outer hair cell efferent innervation in Hoxb1-dependent sensorineural hearing loss. PLoS Genet. 2023;19 (9):e1010933. doi: 10.1371/journal.pgen.1010933. PubMed PMID:37738262 PubMed Central PMC10516434.
- Felske, T, Tocco, C, Péron, S, Harb, K, Alfano, C, Galante, C et al.. Lmo4 synergizes with Fezf2 to promote direct in vivo reprogramming of upper layer cortical neurons and cortical glia towards deep-layer neuron identities. PLoS Biol. 2023;21 (8):e3002237. doi: 10.1371/journal.pbio.3002237. PubMed PMID:37552690 PubMed Central PMC10409279.
- Bonzano, S, Dallorto, E, Molineris, I, Michelon, F, Crisci, I, Gambarotta, G et al.. NR2F1 shapes mitochondria in the mouse brain, providing new insights into Bosch-Boonstra-Schaaf optic atrophy syndrome. Dis Model Mech. 2023;16 (6):. doi: 10.1242/dmm.049854. PubMed PMID:37260288 PubMed Central PMC10309583.
- Studer, M, Rossini, L, Spreafico, R, Pelliccia, V, Tassi, L, de Curtis, M et al.. Why are type II focal cortical dysplasias frequently located at the bottom of sulcus? A neurodevelopmental hypothesis. Epilepsia. 2022;63 (10):2716-2721. doi: 10.1111/epi.17386. PubMed PMID:35932101 .
- Bertacchi, M, Tocco, C, Schaaf, CP, Studer, M. Pathophysiological Heterogeneity of the BBSOA Neurodevelopmental Syndrome. Cells. 2022;11 (8):. doi: 10.3390/cells11081260. PubMed PMID:35455940 PubMed Central PMC9024734.
- Tocco, C, Øvsthus, M, Bjaalie, JG, Leergaard, TB, Studer, M. The topography of corticopontine projections is controlled by postmitotic expression of the area-mapping gene Nr2f1. Development. 2022;149 (5):. doi: 10.1242/dev.200026. PubMed PMID:35262177 PubMed Central PMC8959144.
- Tocco, C, Bertacchi, M, Studer, M. Structural and Functional Aspects of the Neurodevelopmental Gene NR2F1: From Animal Models to Human Pathology. Front Mol Neurosci. 2021;14 :767965. doi: 10.3389/fnmol.2021.767965. PubMed PMID:34975398 PubMed Central PMC8715095.
- Walter, J, Bolognin, S, Poovathingal, SK, Magni, S, Gérard, D, Antony, PMA et al.. The Parkinson's-disease-associated mutation LRRK2-G2019S alters dopaminergic differentiation dynamics via NR2F1. Cell Rep. 2021;37 (3):109864. doi: 10.1016/j.celrep.2021.109864. PubMed PMID:34686322 .
- Jurkute, N, Bertacchi, M, Arno, G, Tocco, C, Kim, US, Kruszewski, AM et al.. Pathogenic NR2F1 variants cause a developmental ocular phenotype recapitulated in a mutant mouse model. Brain Commun. 2021;3 (3):fcab162. doi: 10.1093/braincomms/fcab162. PubMed PMID:34466801 PubMed Central PMC8397830.
- Harb, K, Bertacchi, M, Studer, M. Optimized Immunostaining of Embryonic and Early Postnatal Mouse Brain Sections. Bio Protoc. 2021;11 (1):e3868. doi: 10.21769/BioProtoc.3868. PubMed PMID:33732758 PubMed Central PMC7952938.
2015 - Equipe labélisée - FRM
2011 - Equipe labélisée - FRM
2009 - Senior Chaire d’Excellence - ANR
1997 - MRC (Medical Research Council) Career Development Award (UK)
1994 - EU Fellowship
1993 - EMBO Fellowship

Spotlight on the study published by Michèle Studer in the journal Protein Science in partnership with the “genetic code expansion” (GCE) platform headed by Krittalak Chakrabandhu
Read More

NR2F1, a key factor involved in structural and functional development of the neocortex
Read More
iBV - Institut de Biologie Valrose
"Centre de Biochimie"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
