
Gergo GOGL
Quantitative Interactomics and Disease-Related Networks
Main interests
- Developing innovative quantitative interactomic approaches
- Transient and dynamic protein-protein interactions of cellular signal transduction
- Mapping previously uncharacterized motif-mediated interaction networks
- Understand disease mechanisms based on interactomics
Scientific Questions
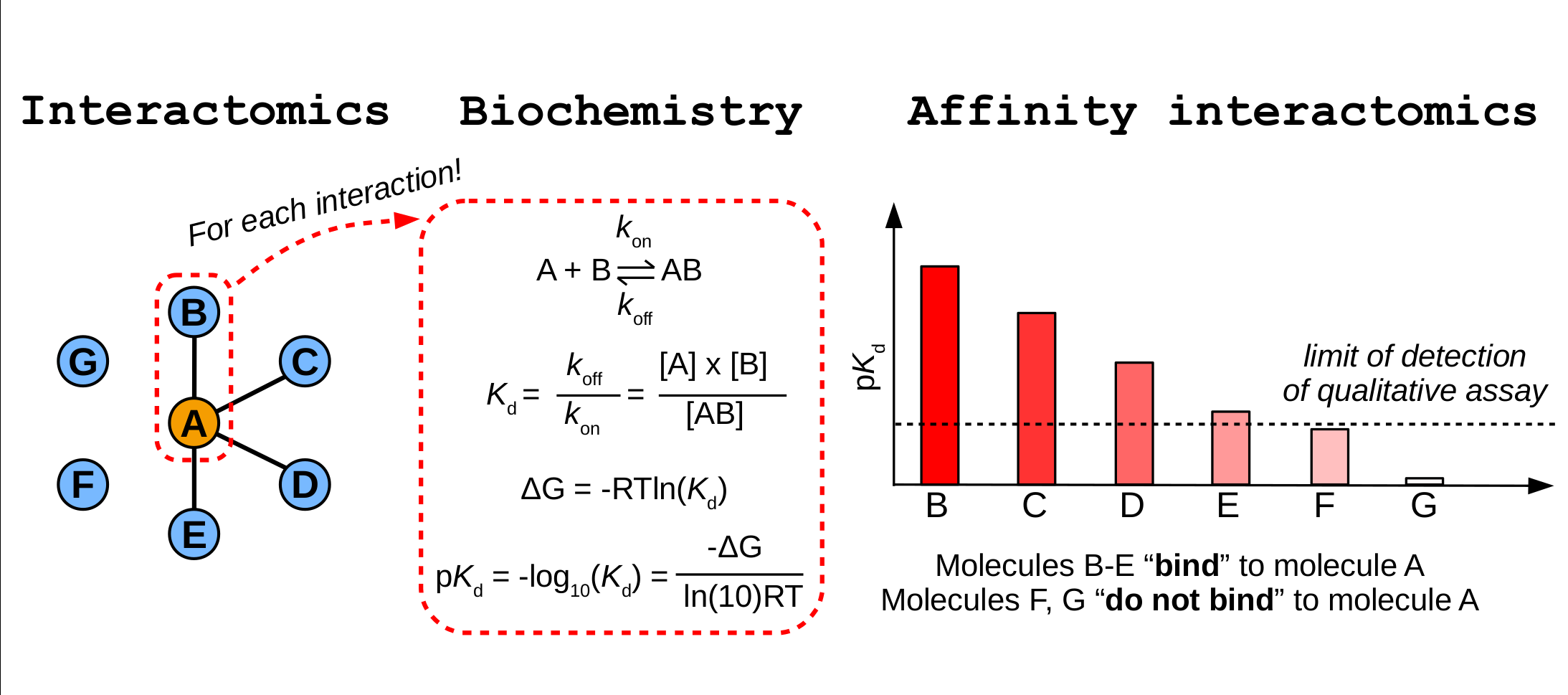
Human cells express 20-25,000 types of proteins. Most of these do not act alone, but they form numerous physical interactions creating functional molecular networks, often referred to as the interactome. These networks are studied by the interactomic field with the ultimate goal to map and investigate interactomes to answer key biological questions. In practice, this is mostly attempted with qualitative approaches, only allowing a glimpse into the overall topology of interaction networks. Our main research objective is to develop alternative, new generation interactomic approaches and to use these tools to answer key biological questions.
We primarily use these approaches to study interactions formed with the help of “short linear interaction motifs”. Motifs are short, but functional peptides found in disordered protein regions that form weak and transient interactions. It is estimated that >80% of all protein-protein interactions are motif-mediated, yet only a small fraction of them are explored experimentally. With the help of new generation quantitative interactomics, we plan to characterize these orphan interactions that are not yet studied experimentally. Such interactions participate in virtually all intracellular signaling processes, such as protein homeostasis, trafficking, or transcription regulation. By uncovering these interactions, we not only understand these specific mechanisms, but we also get a more complete, holistic picture of the whole interactome.
Figure 1 - Illustration of the main differences between the subjects of interactomics, biochemistry, and affinity interactomics.
Our Strategy
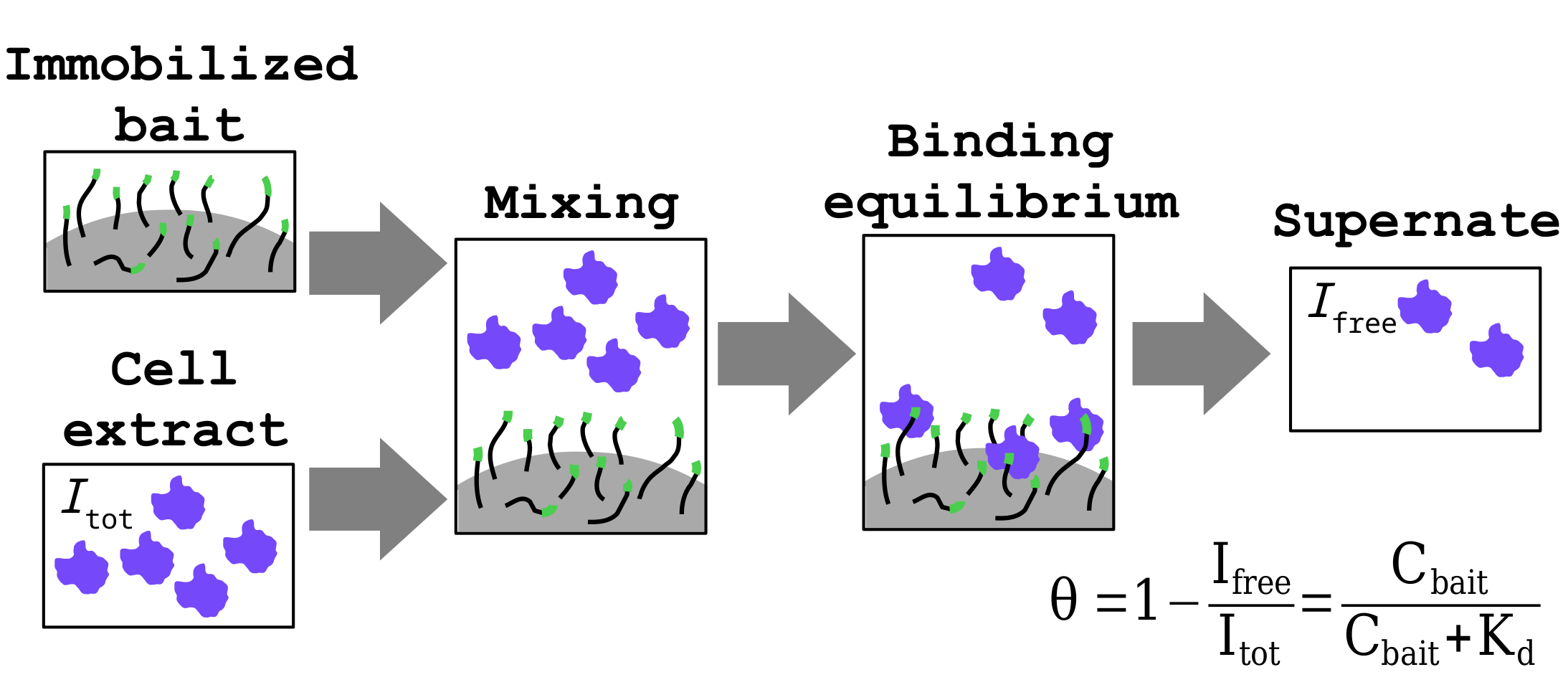
We study molecular networks at two distinct levels. First, we map the intrinsic properties of interactomes by measuring affinities between molecules of our cells. Second, we combine these information with proteomics to estimate how compositions of formed interaction networks changes between different cellular states.
Intrinsic binding affinities are conventionally measured with low-throughput biophysical approaches. These experiments reveal precise and accurate information about binding, such as dissociation constants. In contrast, interactomic approaches are rather qualitative that can only decide if two molecule can or cannot “bind”, with no clear threshold between “binders” and “non-binders”. Our team uses and further develop affinity interactomic approaches, such as the native holdup (nHU) approach, that enable us to measure precise binding affinities on proteomic scales.
Affinities are physical constants and only determined by the nature of the interacting molecules, hence the affinities are practically unchanged between cell types. However, concentrations of the binding partners vary between cells, and even within a single cell, resulting different amounts of formed complexes, or at a system level, different cellular complexomes. We develop ways to combine intrinsic interactomes with these proteomic information to predict these complexomes. We hope that one day, such approach will became a translator that can help us to interpret proteomic changes underlying pathological conditions at a molecular level.
Figure 2 - Schematics of the nHU assay.
Research Aims
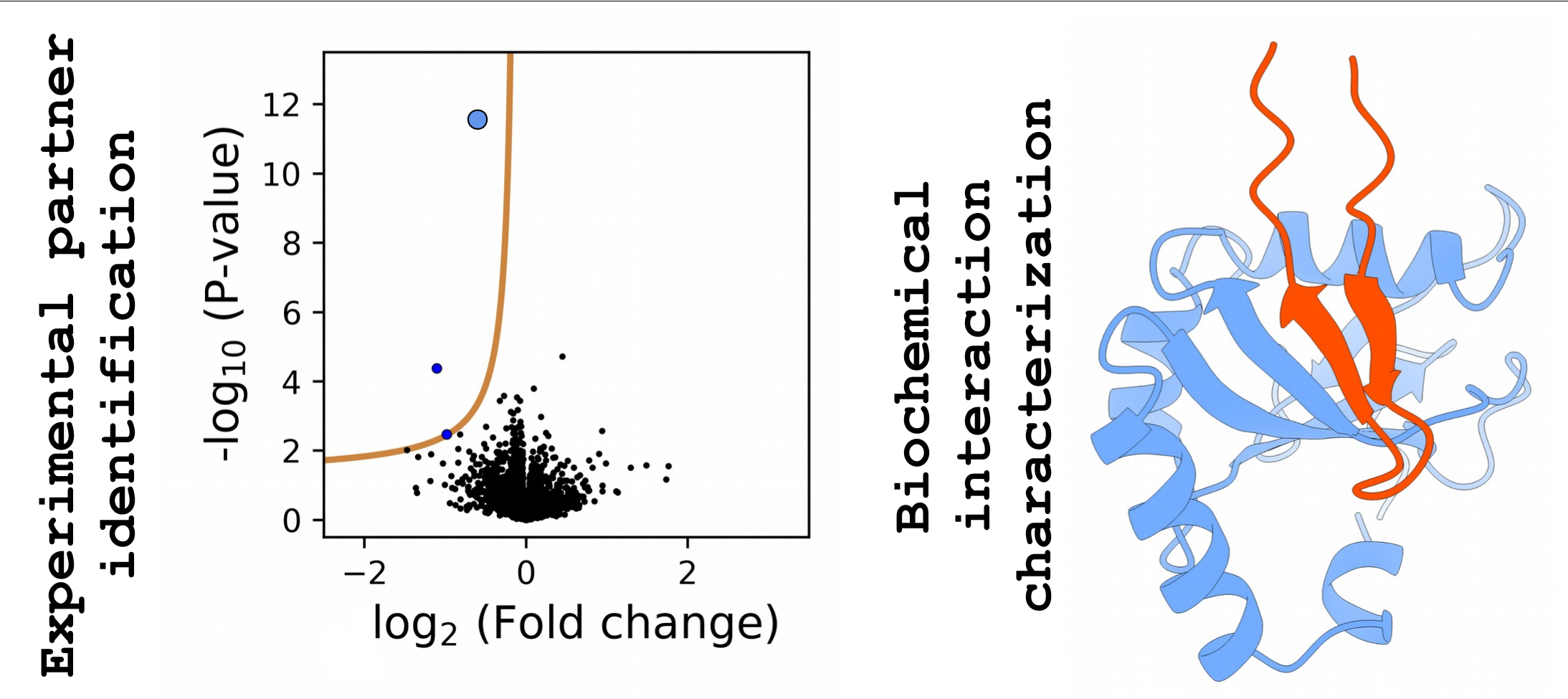
The intrinsic affinity interactome of human proteins, or other molecules, can be mapped, similarly to the human genome. A key aim of our team is to start creating these catalogs of interaction affinities of intact, full-length proteins, or their functional modules. To achieve these goals, we develop cutting edge interactomic approaches, building on a combination of proteomics, biochemistry, and biophysics. This experimental strategy is complemented by computational studies, such as in the form of the ProfAff affinity interactomic web database (for Profiling Affinities) that is developed by our team.
Figure 3 - A proteome-wide nHU experiment with its identified interaction visualized by an AlphaFold prediction.
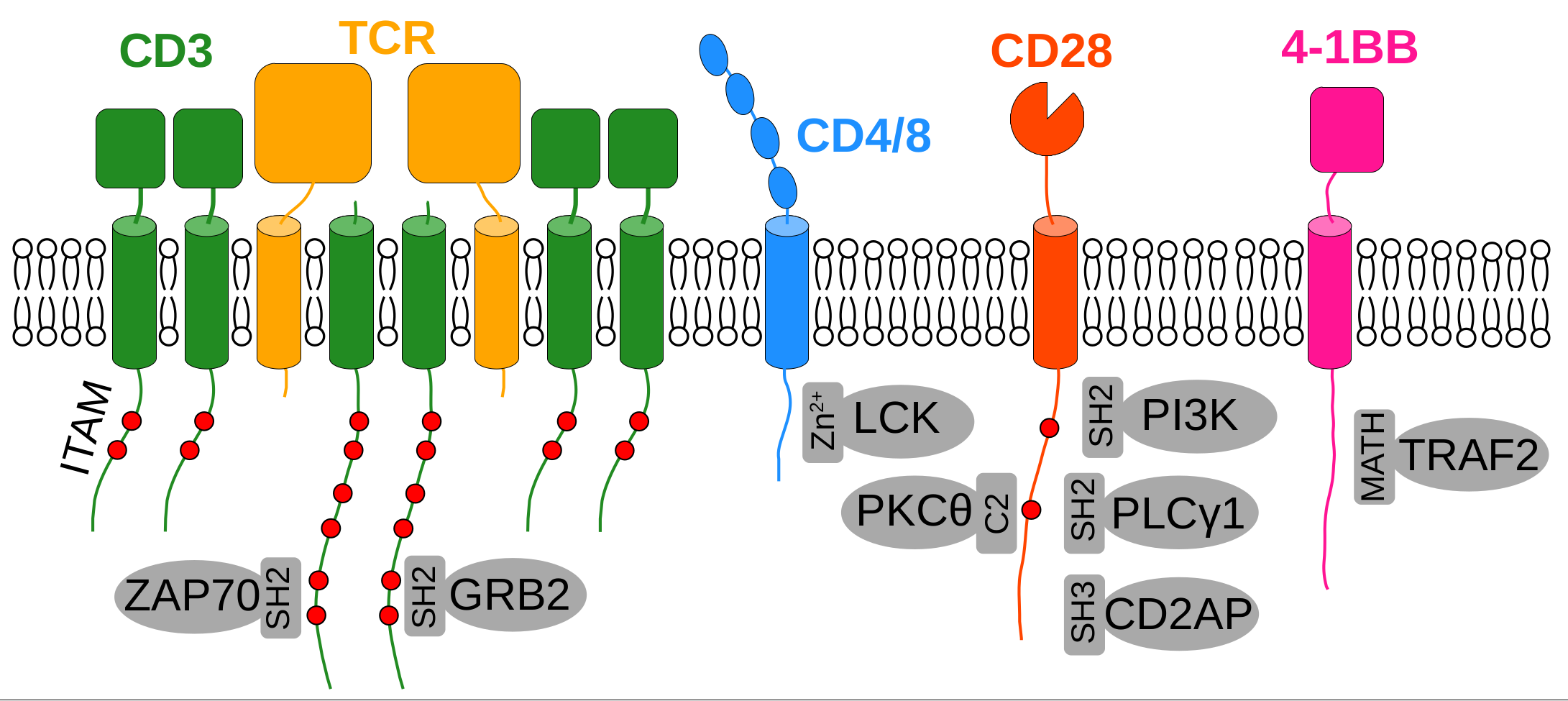
Our proteome contains thousands of modular domains that can interact with peptide motifs and these interactions are the fundamental building blocks of many processes, such as intracellular signal transduction. Motif-mediated interactions are typically promiscuous, where a single domain can recognize hundreds of motifs and a single motif can recognize dozens of protein domains. They are also typically weak and transient, allowing fast exchange between bound and unbound molecules, forming highly dynamic networks. To this day, only a small fraction of these motif-mediated networks have been mapped and our group specifically focuses on orphan motifs that do not yet have a recognized interaction partners but can be identified based on evolutionary or functional data, such as recurring disease-related mutations or post-translational modifications.
Figure 4 - Selected motif-mediated interactions underpinning immune signaling.
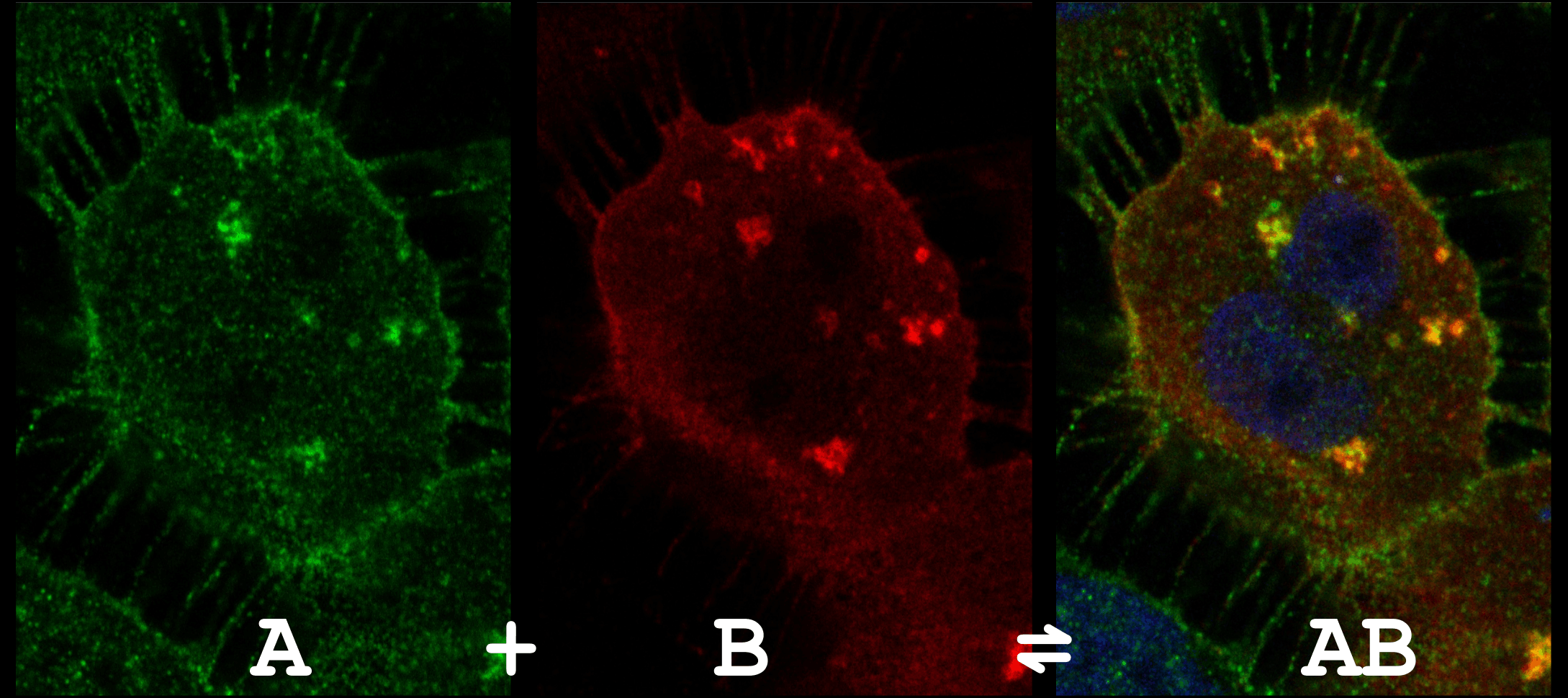
Many biological processes, like signal transduction, is mediated by a large number of transient interactions. These interactions have weak intrinsic affinities and are often regulated dynamically by post-translational modifications. Not surprisingly, it is highly challenging to capture these interactions in cells where they form complexes at low amounts with very short lifetimes. Yet, these low abundant interactions mediate a multitude of cellular mechanisms. Our group is using various types of protein-protein interaction sensors to trap and visualize these macromolecular interactions in situ and to investigate the roles of these complexes in specific cellular processes.
Figure 5 - A co-localization assay of a protein-protein interaction.
Researchers
 JEZSO Balint - +33 489153717
JEZSO Balint - +33 489153717 KIRIGIN Elisa - +33 R
KIRIGIN Elisa - +33 R
Postdocs
 ZAMBO Boglarka - +33 489153717
ZAMBO Boglarka - +33 489153717 ZAMBO Boglarka - +33 489153717
ZAMBO Boglarka - +33 489153717
Students
Recent publications
- Keskitalo, S, Zambo, B, Malaymar Pinar, D, Tuhkala, A, Salokas, K, Turunen, T et al.. The non-catalytic DNA polymerase ε subunit is an NPF motif recognition protein. Nat Commun. 2025;17 (1):586. doi: 10.1038/s41467-025-67284-5. PubMed PMID:41390769 PubMed Central PMC12808100.
- Delalande, F, Østergaard, SR, Gogl, G, Cousido-Siah, A, McEwen, AG, Men, Y et al.. Holdup Multiplex Assay for High-Throughput Measurement of Protein-Ligand Affinity Constants Using a Mass Spectrometry Readout. J Am Chem Soc. 2025;147 (13):10886-10902. doi: 10.1021/jacs.4c11102. PubMed PMID:40129024 .
- Póti, ÁL, Bálint, D, Alexa, A, Sok, P, Ozsváth, K, Albert, K et al.. Targeting a key protein-protein interaction surface on mitogen-activated protein kinases by a precision-guided warhead scaffold. Nat Commun. 2024;15 (1):8607. doi: 10.1038/s41467-024-52574-1. PubMed PMID:39366929 PubMed Central PMC11452651.
- Zambo, B, Edelweiss, E, Morlet, B, Negroni, L, Pajkos, M, Dosztanyi, Z et al.. Uncovering the BIN1-SH3 interactome underpinning centronuclear myopathy. Elife. 2024;13 :. doi: 10.7554/eLife.95397. PubMed PMID:38995680 PubMed Central PMC11245310.
- Jimenez-Armijo, A, Morkmued, S, Ahumada, JT, Kharouf, N, de Feraudy, Y, Gogl, G et al.. The Rogdi knockout mouse is a model for Kohlschütter-Tönz syndrome. Sci Rep. 2024;14 (1):445. doi: 10.1038/s41598-023-50870-2. PubMed PMID:38172607 PubMed Central PMC10764811.
- Montserrat-Gomez, M, Gogl, G, Carrasco, K, Betzi, S, Durbesson, F, Cousido-Siah, A et al.. PDZome-wide and structural characterization of the PDZ-binding motif of VANGL2. Biochim Biophys Acta Proteins Proteom. 2024;1872 (3):140989. doi: 10.1016/j.bbapap.2023.140989. PubMed PMID:38142947 .
- Zambo, B, Gogl, G, Morlet, B, Eberling, P, Negroni, L, Moine, H et al.. Comparative analysis of PDZ-binding motifs in the diacylglycerol kinase family. FEBS J. 2024;291 (4):690-704. doi: 10.1111/febs.16994. PubMed PMID:37942667 .
- Weimer, K, Zambo, B, Gogl, G. Molecules interact. But how strong and how much?. Bioessays. 2023;45 (6):e2300007. doi: 10.1002/bies.202300007. PubMed PMID:36998104 .
- Zambo, B, Morlet, B, Negroni, L, Trave, G, Gogl, G. Native holdup (nHU) to measure binding affinities from cell extracts. Sci Adv. 2022;8 (51):eade3828. doi: 10.1126/sciadv.ade3828. PubMed PMID:36542723 PubMed Central PMC9770967.
- Gogl, G, Zambo, B, Kostmann, C, Cousido-Siah, A, Morlet, B, Durbesson, F et al.. Author Correction: Quantitative fragmentomics allow affinity mapping of interactomes. Nat Commun. 2022;13 (1):7555. doi: 10.1038/s41467-022-35177-6. PubMed PMID:36477203 PubMed Central PMC9729566.
- Gogl, G, Zambo, B, Kostmann, C, Cousido-Siah, A, Morlet, B, Durbesson, F et al.. Quantitative fragmentomics allow affinity mapping of interactomes. Nat Commun. 2022;13 (1):5472. doi: 10.1038/s41467-022-33018-0. PubMed PMID:36115835 PubMed Central PMC9482650.
- Simon, MA, Bartus, É, Mag, B, Boros, E, Roszjár, L, Gógl, G et al.. Promiscuity mapping of the S100 protein family using a high-throughput holdup assay. Sci Rep. 2022;12 (1):5904. doi: 10.1038/s41598-022-09574-2. PubMed PMID:35393447 PubMed Central PMC8991199.
- Cousido-Siah, A, Carneiro, L, Kostmann, C, Ecsedi, P, Nyitray, L, Trave, G et al.. A scalable strategy to solve structures of PDZ domains and their complexes. Acta Crystallogr D Struct Biol. 2022;78 (Pt 4):509-516. doi: 10.1107/S2059798322001784. PubMed PMID:35362473 .
- Alexa, A, Sok, P, Gross, F, Albert, K, Kobori, E, Póti, ÁL et al.. A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host. Nat Commun. 2022;13 (1):472. doi: 10.1038/s41467-022-28109-x. PubMed PMID:35078976 PubMed Central PMC8789800.
- Ecsédi, P, Gógl, G, Nyitray, L. Studying the Structures of Relaxed and Fuzzy Interactions: The Diverse World of S100 Complexes. Front Mol Biosci. 2021;8 :749052. doi: 10.3389/fmolb.2021.749052. PubMed PMID:34708078 PubMed Central PMC8542695.
- Merő, B, Koprivanacz, K, Cserkaszky, A, Radnai, L, Vas, V, Kudlik, G et al.. Characterization of the Intramolecular Interactions and Regulatory Mechanisms of the Scaffold Protein Tks4. Int J Mol Sci. 2021;22 (15):. doi: 10.3390/ijms22158103. PubMed PMID:34360869 PubMed Central PMC8348221.
- Bauer, V, Schmidtgall, B, Gógl, G, Dolenc, J, Osz, J, Nominé, Y et al.. Conformational editing of intrinsically disordered protein by α-methylation. Chem Sci. 2020;12 (3):1080-1089. doi: 10.1039/d0sc04482b. PubMed PMID:34163874 PubMed Central PMC8178997.
- Caillet-Saguy, C, Durbesson, F, Rezelj, VV, Gogl, G, Tran, QD, Twizere, JC et al.. Host PDZ-containing proteins targeted by SARS-CoV-2. FEBS J. 2021;288 (17):5148-5162. doi: 10.1111/febs.15881. PubMed PMID:33864728 PubMed Central PMC8250131.
- Gogl, G, Tugaeva, KV, Eberling, P, Kostmann, C, Trave, G, Sluchanko, NN et al.. Hierarchized phosphotarget binding by the seven human 14-3-3 isoforms. Nat Commun. 2021;12 (1):1677. doi: 10.1038/s41467-021-21908-8. PubMed PMID:33723253 PubMed Central PMC7961048.
- Jané, P, Gógl, G, Kostmann, C, Bich, G, Girault, V, Caillet-Saguy, C et al.. Interactomic affinity profiling by holdup assay: Acetylation and distal residues impact the PDZome-binding specificity of PTEN phosphatase. PLoS One. 2020;15 (12):e0244613. doi: 10.1371/journal.pone.0244613. PubMed PMID:33382810 PubMed Central PMC7774954.
2024 - ATIP-Avenir Junior Group Leader Program

Gergő Gógl, laureate of the 2025 Impulscience® program for his work on transient protein interactions
Read More
iBV - Institut de Biologie Valrose
"Tour Pasteur"
Université Nice Sophia Antipolis
Faculté de médecine
28 Avenue de Valombrose
06189 Nice cedex 2
