
Xavier DESCOMBES
Morphology and Image (MORPHEME)
Research Axes
- Imaging
- Feature extraction
- Interpretation/Classification
- Modeling
Scientific Questions
We aim to develop new numerical tools for analyzing bio-images. The targeted biological structures go from the sub-cellular to the tissue scale. The addressed problems concern the image reconstruction and restoration, objects detection, machine learning and modeling. Our methods include inverse problems, geometric and stochastic approaches and deep learning. We address data of different sources including fluorescence and reflectance images or histopathological WSI data. Our results concern small particles (such as RNA granules) detection and characterization, cells, nuclei detection, fiber networks extraction and characterization, tissue classification, urchin morphogenesis and plankton quantification.
Our Strategy
Our work is built on a multidisciplinary framework including specialists in image processing, biophysics, machine learning and biology. Our strategy is to embed biological properties or prior into numerical and mathematical framework. To this end, we collaborate with biologists of several teams to co-construct numerical solutions for reconstructing, extracting, classifying and modeling the information contained in images. We jointly evaluate existing software and consequently develop new approaches to overcome their limits. We propose software that are open source, mostly coded in Python.
Research Axes
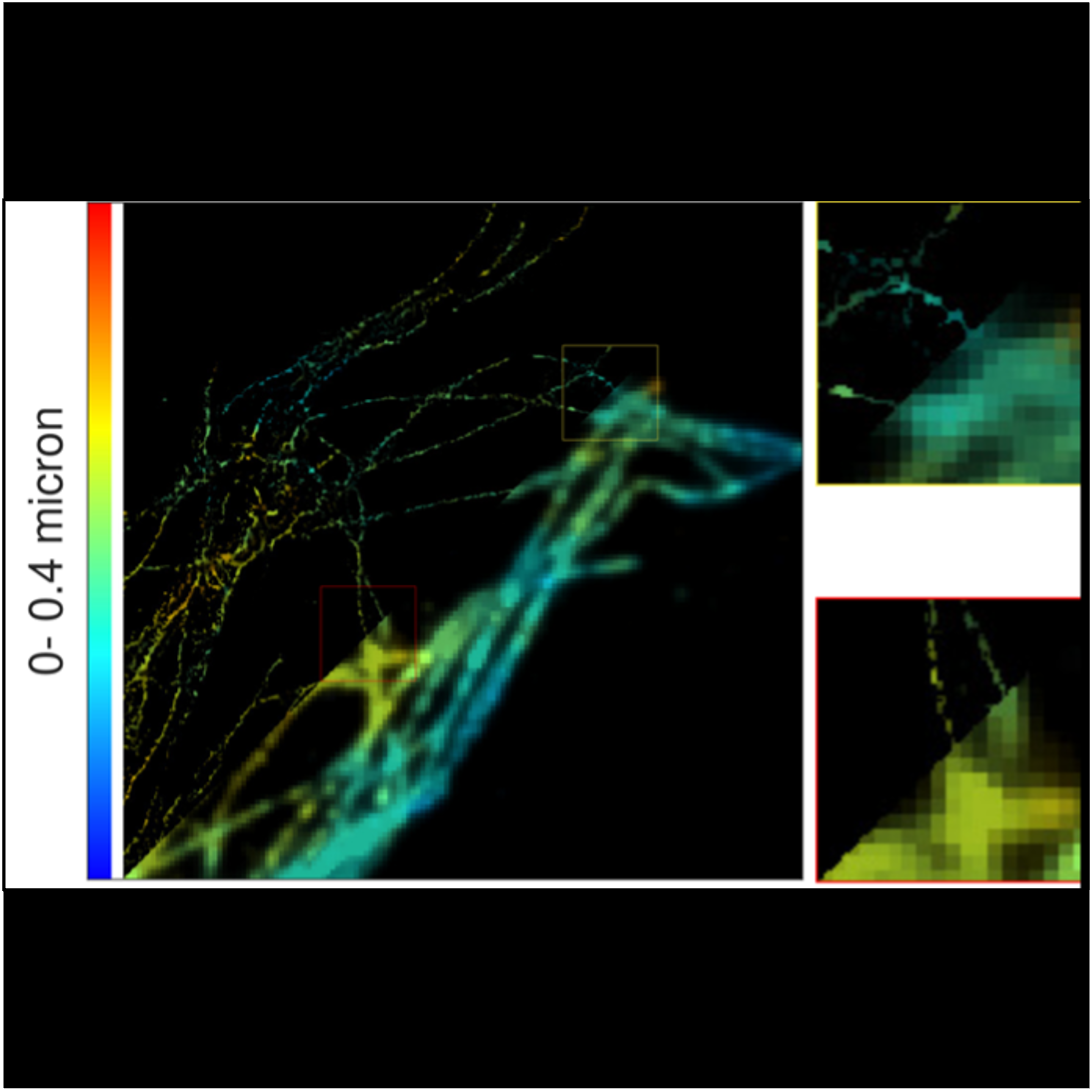
Image reconstruction
Our aim is to produce the best image quality, with high resolution and no noise, from fluorescence microscopes. We focus on super-resolution reconstruction in the 3D domain, from SMLM data or molecule fluctuations in the transverse plane. Super-resolution by fluctuation of molecule does not need specific microscope nor fluorophore. By modeling the physics of acquisition, we build algorithms that reconstruct images well resolved with less noise from observations with specific geometry, as in TIRF-MA for example.
Figure: Super-resolution reconstruction of a TIRF image
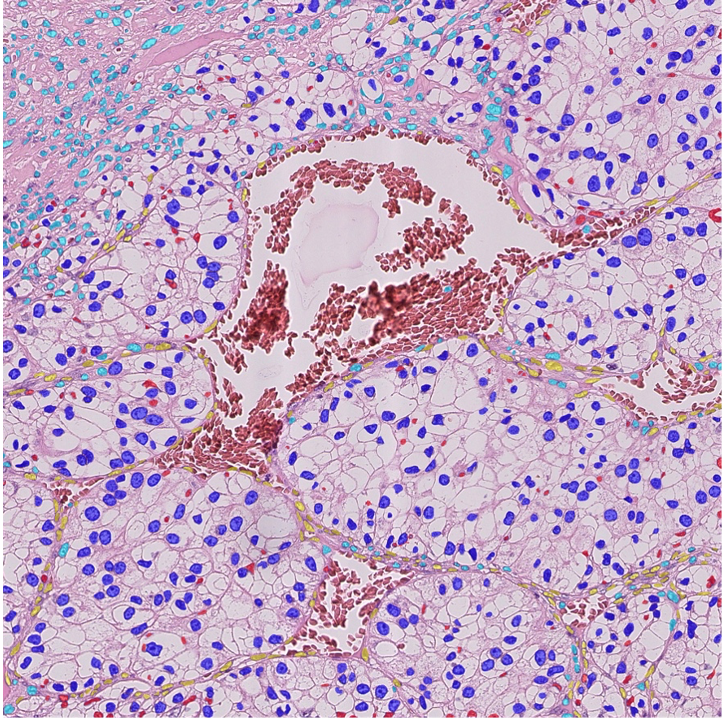
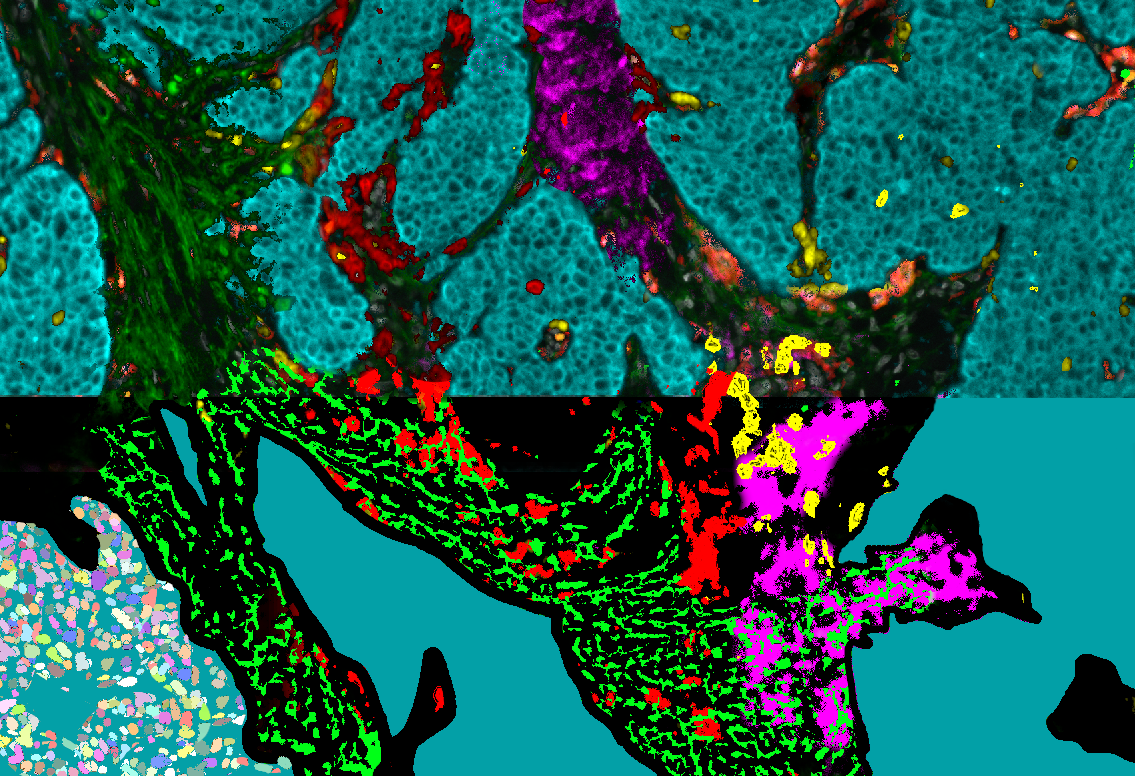
Object detection
A preliminary step to analyze a tissue is to detect and segment its components at different scales. This includes vessels, cell nuclei, intra-cellular particles. We are investigating two main methodological axes based on stochastic models (marked point processes) that embed geometrical properties of the targeted objects (SPADE, ObjMPP software) and on deep learning technics based on annotated training database. Applications include RNA-IMP granules (coll. With F. De Graeve, F. Besse, IBV) and histopathology (coll. with D. Ambrosetti, CHU Nice, N. Pote Bichat Hospital).
Figure: Nuclei detection and classification from HE histological images

Cell lineage analysis
Microscopy techniques allow to image temporal 3D stacks of developing organs or embryos at the cellular level and with an acquisition frequency that enables to retrieve not only the individual cell morphology but also the cell lineages. Imaging a few organs or embryos with several conditions may help to decipher the various components that drive the development. This necessitates to distinguish the inter-individual variability from the components’ effects. We are currently working on two models, ascidian embryos within a collaboration with P. Lemaire (CRBM, Montpellier) and urchin embryos with M. Rauzi (IBV, Nice).
Figure: Cell lineage in urchin embryo
2023 - Laure Blanc-Féraud has been awarded of the CNRS silver medal
https://www.ins2i.cnrs.fr/fr/cnrsinfo/laure-blanc-feraud-repousser-les-frontieres-de-limagerie.
2023 - Luca Calatroni has been awarded an ERC Starting Grant
https://www.ins2i.cnrs.fr/fr/cnrsinfo/erc-starting-grant-luca-calatroni-et-la-quete-de-la-super-resolution-par-apprentissage
Laboratoire d’Informatique, Signaux et Systèmes de Sophia-Antipolis (I3S)
Les Algorithmes – bât. Euclide B
2000 Route Des Lucioles
06900 Sophia Antipolis
France
iBV - Institut de Biologie Valrose
"Centre de Biochimie"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2




