
Franck DELAUNAY
Chronobiology
Main interests
- Circadian clock
- Metabolism
- Cell cycle
- Systems biology
Scientific Questions

The mammalian circadian timing system coordinates a variety of molecular, cellular and physiological processes over the 24 h in synchrony with the light/dark cycle. The fundamental mechanism underlying circadian rhythms is a molecular clock present in virtually all body cells. The system is organized hierarchically with a central clock in the hypothalamus that coordinates peripheral clocks through internal synchronizers. Misalignment or disruption of the human circadian timing system has been linked to pathologies including metabolic disorders, sleep phase syndromes and cancer. Although we currently have a comprehensive view of how core clock genes work and interact together to generate oscillations, we still do not fully understand how they crosstalk with other essential cellular mechanisms. In that context, we study how mammalian peripheral circadian clocks interact with the cell cycle and metabolic processes and determine the relevance of these interactions in health and disease.
Our Strategy
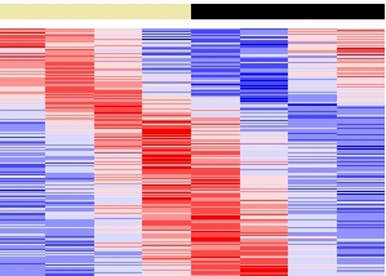
To study the interaction between the circadian clock and metabolism, we use a panel of tools and approaches including systemic and conditional knockout mouse models, culture of primary cells, functional genomics, nutritional challenges and, monitoring of circadian physiology and behavior. Our second project on the interaction between the circadian clock and the cell cycle involves mainly a systems biology approach that combines multispectral fluorescent reporter systems, high throughput time lapse imaging, bioinformatics and mathematical modelling in collaboration with mathematicians and computer scientists.
Research Aims
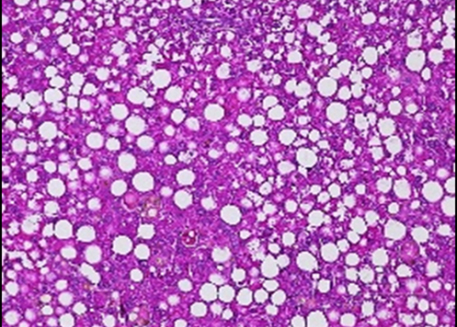
We have established that Krüppel Like Factor 10 (KLF10) is a clock-controlled transcription factor in liver. Our goal is to precisely decipher its role in the circadian coordination of key metabolic processes in hepatocytes. We also aim to evaluate its relevance in the context of non-alcoholic fatty liver disease which is rising worldwide.
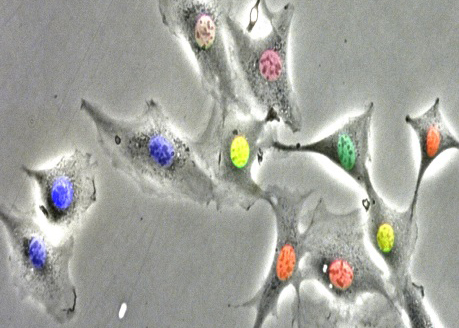
We have demonstrated that the two oscillators driving progression of the cell cycle and the circadian clock respectively are robustly coupled through a phase locking mechanism in unperturbed dividing mammalian cells. Our main goal is to understand the dynamics of this coupling and determine how this phenomenon might be exploited to design chronotherapeutics approaches for cancer treatment.
Researchers
 GRECHEZ-CASSIAU Aline - +33 489150826
GRECHEZ-CASSIAU Aline - +33 489150826 TEBOUL Michèle - +33 489150825
TEBOUL Michèle - +33 489150825
PreDocs
 MARCHANDISE-FRANQUET Sophia - +33 R
MARCHANDISE-FRANQUET Sophia - +33 R
Engineers & Technicians
 GUéRIN Sophie - +33 489150827
GUéRIN Sophie - +33 489150827 KRAWCZYK Inès - +33 R
KRAWCZYK Inès - +33 R TAHRAOUI Sana - +33 R
TAHRAOUI Sana - +33 R
Students
 SAHLI Wiem - +33 R
SAHLI Wiem - +33 R
Recent Publications
- Burckard, O, Teboul, M, Delaunay, F, Chaves, M. Benchmark for quantitative characterization of circadian clock cycles. Biosystems. 2025;247 :105363. doi: 10.1016/j.biosystems.2024.105363. PubMed PMID:39551427 .
- Teboul, M, Delaunay, F. [Harmonic oscillations of circadian rhythms come out of the shadows]. Med Sci (Paris). 2023;39 (6-7):544-550. doi: 10.1051/medsci/2023079. PubMed PMID:37387663 .
- Burckard, O, Teboul, M, Delaunay, F, Chaves, M. Cycle dynamics and synchronization in a coupled network of peripheral circadian clocks. Interface Focus. 2022;12 (3):20210087. doi: 10.1098/rsfs.2021.0087. PubMed PMID:35464139 PubMed Central PMC9010852.
- Ruberto, AA, Gréchez-Cassiau, A, Guérin, S, Martin, L, Revel, JS, Mehiri, M et al.. KLF10 integrates circadian timing and sugar signaling to coordinate hepatic metabolism. Elife. 2021;10 :. doi: 10.7554/eLife.65574. PubMed PMID:34402428 PubMed Central PMC8410083.
- Rougny, A, Paulevé, L, Teboul, M, Delaunay, F. A detailed map of coupled circadian clock and cell cycle with qualitative dynamics validation. BMC Bioinformatics. 2021;22 (1):240. doi: 10.1186/s12859-021-04158-9. PubMed PMID:33975535 PubMed Central PMC8114686.
- Leclère, PS, Rousseau, D, Patouraux, S, Guérin, S, Bonnafous, S, Gréchez-Cassiau, A et al.. MCD diet-induced steatohepatitis generates a diurnal rhythm of associated biomarkers and worsens liver injury in Klf10 deficient mice. Sci Rep. 2020;10 (1):12139. doi: 10.1038/s41598-020-69085-w. PubMed PMID:32699233 PubMed Central PMC7376252.
- Almeida, S, Chaves, M, Delaunay, F. Transcription-based circadian mechanism controls the duration of molecular clock states in response to signaling inputs. J Theor Biol. 2020;484 :110015. doi: 10.1016/j.jtbi.2019.110015. PubMed PMID:31539528 .
- Mura, M, Feillet, C, Bertolusso, R, Delaunay, F, Kimmel, M. Mathematical modelling reveals unexpected inheritance and variability patterns of cell cycle parameters in mammalian cells. PLoS Comput Biol. 2019;15 (6):e1007054. doi: 10.1371/journal.pcbi.1007054. PubMed PMID:31158226 PubMed Central PMC6564046.
- Canaple, L, Gréchez-Cassiau, A, Delaunay, F, Dkhissi-Benyahya, O, Samarut, J. Maternal eating behavior is a major synchronizer of fetal and postnatal peripheral clocks in mice. Cell Mol Life Sci. 2018;75 (21):3991-4005. doi: 10.1007/s00018-018-2845-5. PubMed PMID:29804258 PubMed Central PMC11105238.
- Teboul, M, Delaunay, F. [Biological clocks: a rythm can hide another one]. Med Sci (Paris). 2018;34 (1):30-33. doi: 10.1051/medsci/20183401009. PubMed PMID:29384092 .
- Delaunay, F. [The 2017 Nobel prize in physiology and medicine for the circardian clock]. Bull Cancer. 2017;104 (10):821-822. doi: 10.1016/j.bulcan.2017.10.005. PubMed PMID:29107314 .
- Traynard, P, Feillet, C, Soliman, S, Delaunay, F, Fages, F. Model-based investigation of the circadian clock and cell cycle coupling in mouse embryonic fibroblasts: Prediction of RevErb-α up-regulation during mitosis. Biosystems. 2016;149 :59-69. doi: 10.1016/j.biosystems.2016.07.003. PubMed PMID:27443484 .
- Feillet, C, Guérin, S, Lonchampt, M, Dacquet, C, Gustafsson, JÅ, Delaunay, F et al.. Sexual Dimorphism in Circadian Physiology Is Altered in LXRα Deficient Mice. PLoS One. 2016;11 (3):e0150665. doi: 10.1371/journal.pone.0150665. PubMed PMID:26938655 PubMed Central PMC4777295.
- Behaegel, J, Comet, JP, Bernot, G, Cornillon, E, Delaunay, F. A hybrid model of cell cycle in mammals. J Bioinform Comput Biol. 2016;14 (1):1640001. doi: 10.1142/S0219720016400011. PubMed PMID:26708052 .
- Gréchez-Cassiau, A, Feillet, C, Guérin, S, Delaunay, F. The hepatic circadian clock regulates the choline kinase α gene through the BMAL1-REV-ERBα axis. Chronobiol Int. 2015;32 (6):774-84. doi: 10.3109/07420528.2015.1046601. PubMed PMID:26125130 .
- Feillet, C, van der Horst, GT, Levi, F, Rand, DA, Delaunay, F. Coupling between the Circadian Clock and Cell Cycle Oscillators: Implication for Healthy Cells and Malignant Growth. Front Neurol. 2015;6 :96. doi: 10.3389/fneur.2015.00096. PubMed PMID:26029155 PubMed Central PMC4426821.
- Feillet, C, Krusche, P, Tamanini, F, Janssens, RC, Downey, MJ, Martin, P et al.. Phase locking and multiple oscillating attractors for the coupled mammalian clock and cell cycle. Proc Natl Acad Sci U S A. 2014;111 (27):9828-33. doi: 10.1073/pnas.1320474111. PubMed PMID:24958884 PubMed Central PMC4103330.
- Siffroi-Fernandez, S, Dulong, S, Li, XM, Filipski, E, Gréchez-Cassiau, A, Peteri-Brünback, B et al.. Functional genomics identify Birc5/survivin as a candidate gene involved in the chronotoxicity of cyclin-dependent kinase inhibitors. Cell Cycle. 2014;13 (6):984-91. doi: 10.4161/cc.27868. PubMed PMID:24552823 PubMed Central PMC3984321.
- Li, XM, Mohammad-Djafari, A, Dumitru, M, Dulong, S, Filipski, E, Siffroi-Fernandez, S et al.. A circadian clock transcription model for the personalization of cancer chronotherapy. Cancer Res. 2013;73 (24):7176-88. doi: 10.1158/0008-5472.CAN-13-1528. PubMed PMID:24154875 .
- Billy, F, Clairambault, J, Delaunay, F, Feillet, C, Robert, N. Age-structured cell population model to study the influence of growth factors on cell cycle dynamics. Math Biosci Eng. 2013;10 (1):1-17. doi: 10.3934/mbe.2013.10.1. PubMed PMID:23311359 .
iBV - Institut de Biologie Valrose
"Sciences Naturelles"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2
