
Agnès BANRETI
CHIRALIFE: Homochirality Regulation and the Role of Heterochirality in Aging and Disease
Main interests
- Investigating the underlying mechanisms and conditions where loss of homochirality contributes to physiopathological conditions.
- Identifying evolutionary conserved epimerization hotspots across the proteome.
- Uncovering a novel family of genes responsible for maintaining biological homochirality.
- Developing model-independent state-of-the-art interdisciplinary methodologies to study dynamic chirality changes in vitro and in vivo.
Scientific Questions
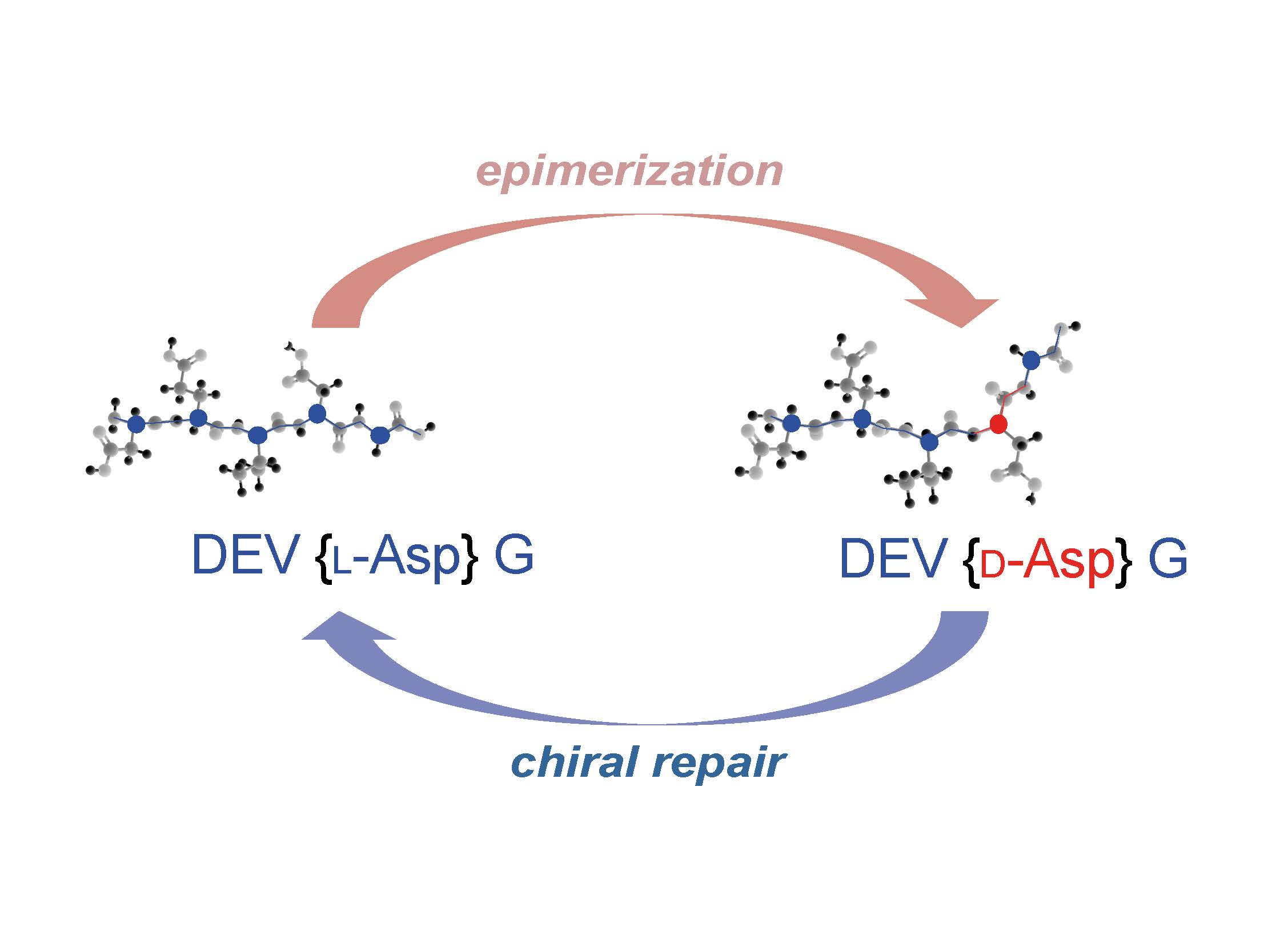
One of the most fascinating and unexplored questions in the life sciences is the origin and significance of biomolecular homochirality, a key feature that distinguishes all living organisms from inorganic/abiotic matter. Our interdisciplinary team aims to understand how organisms actively maintain the homochirality of their proteins. Specifically, we focus on identifying genes and mechanisms that regulate chirality by repairing post-translationally epimerized amino acid residues in proteins or eliminating heterochiral proteins. Our integrative research also seeks to uncover the mechanisms behind the direct pathological consequences of losing proteome homochirality in vivo.
Figure 1. Enzymatic repair of post-translationally epimerized residues ensures the maintenance of protein homochirality.
Our Strategy
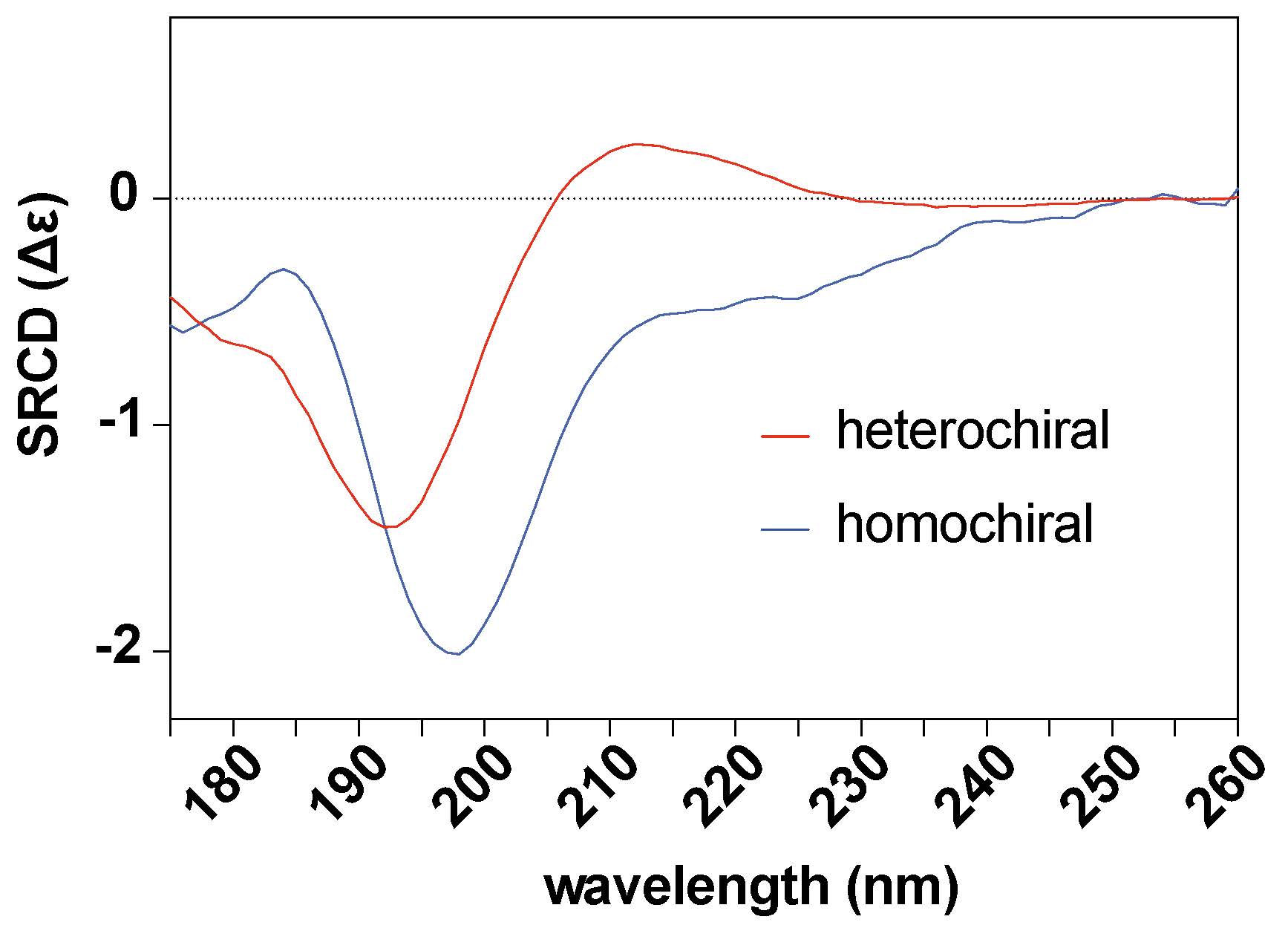
We have developed various methods and tools to monitor chirality changes and identify proteins with non-L-amino acids formed post-translationally through epimerization or isomerization in vivo, using Drosophila as a pilot model system. To understand the physiological relevance of homochirality regulation, we experimentally enhance heterochirality and investigate the molecular mechanisms underlying the physiological consequences of accumulating non-L-amino acid-containing proteins (Aim 1). We aim to map epimerization sites in evolutionarily conserved proteins using chiral-selective large-scale proteomics, bioinformatics, and AI-driven machine learning approaches (Aim 2). To understand how proteome L-homochirality is maintained in living organisms, we combine interdisciplinary chiral-selective techniques (analytical chemistry, physicochemistry) with genetic screening to identify chirality-regulating genes dedicated to preserving homochirality (Aim 3).
Figure 2. Far-UV Synchrotron Radiation Circular Dichroism (SRCD) spectra of homochiral and heterochiral pentamer peptides that were used as epitopes for chiral-selective antibody generation.
Research Aims
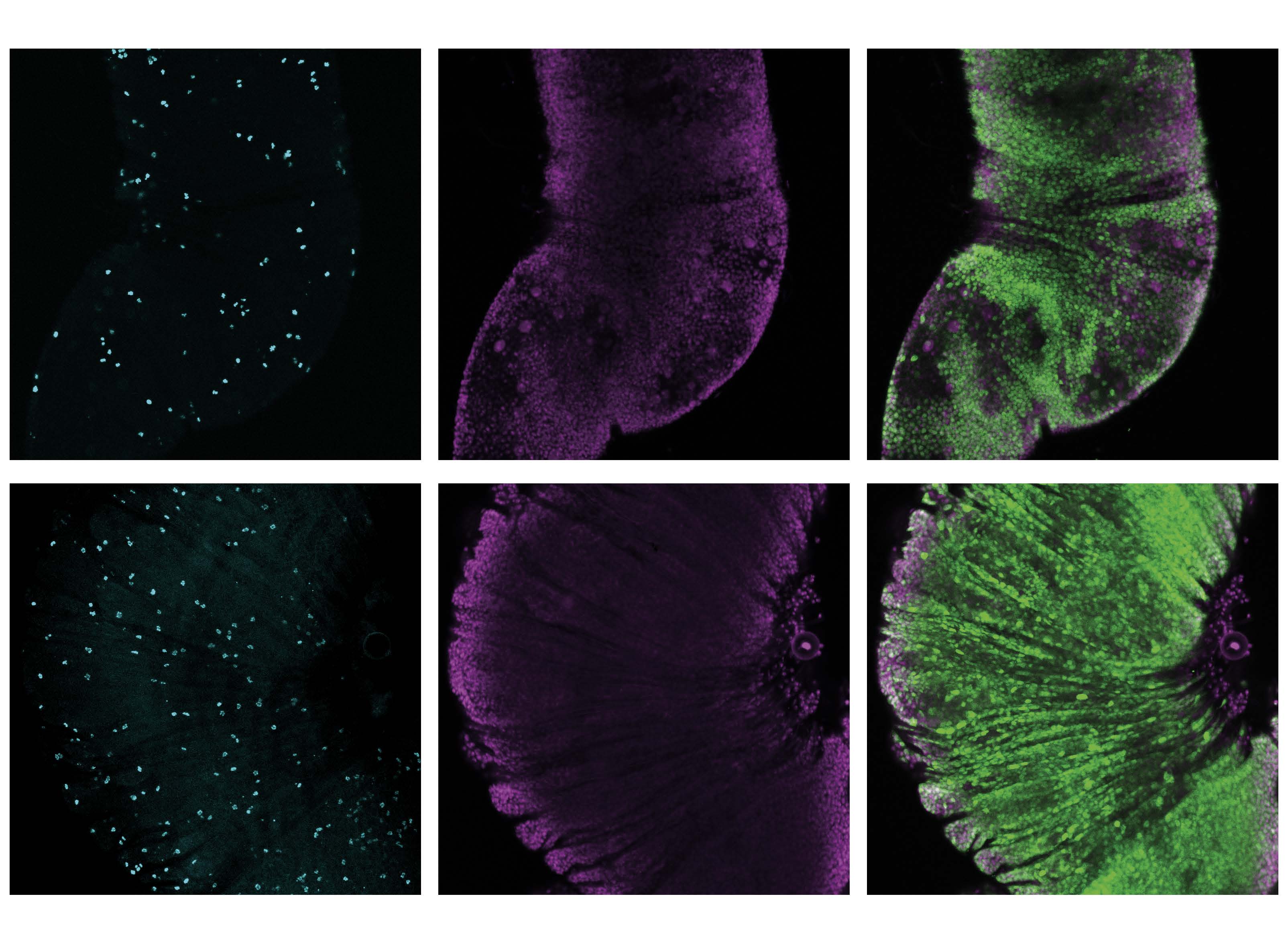
Our research established a direct link between D-amino acids and protein dysfunction in vivo, leading to a progressive "heterochirality syndrome." This syndrome cascades across biological scales, from the loss of molecular homochirality to increased tumor susceptibility in organs, ultimately resulting in a shortened lifespan in chiral-deficient animals. We aim to deepen our understanding of the molecular, cellular, and developmental mechanisms that connect heterochirality to these biological effects, employing a range of genetic, cellular, and biochemical approaches.
Figure 3. Intestinal tumor (green) in a chiral-deficient animal.
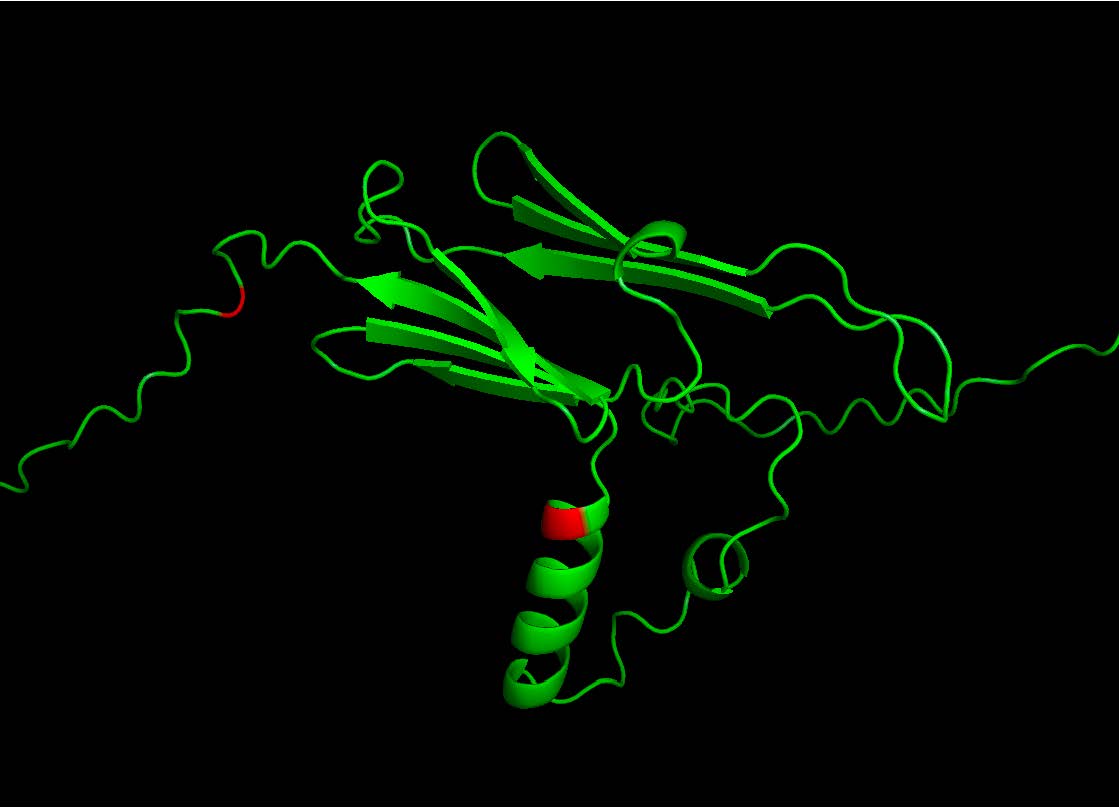
Our goal is to pinpoint hotspot sequences most prone to epimerization and expand our studies to various chiral amino acids. Identifying new heterochiral proteins will help us explore novel cellular and physiological processes affected by heterochirality. In the long term, D-neoepitopes identified in the proteome could serve as targets, enabling the design of drugs that selectively target proteins containing D-amino acids.
Figure 4. Identified epimerization hot spots are marked in red.
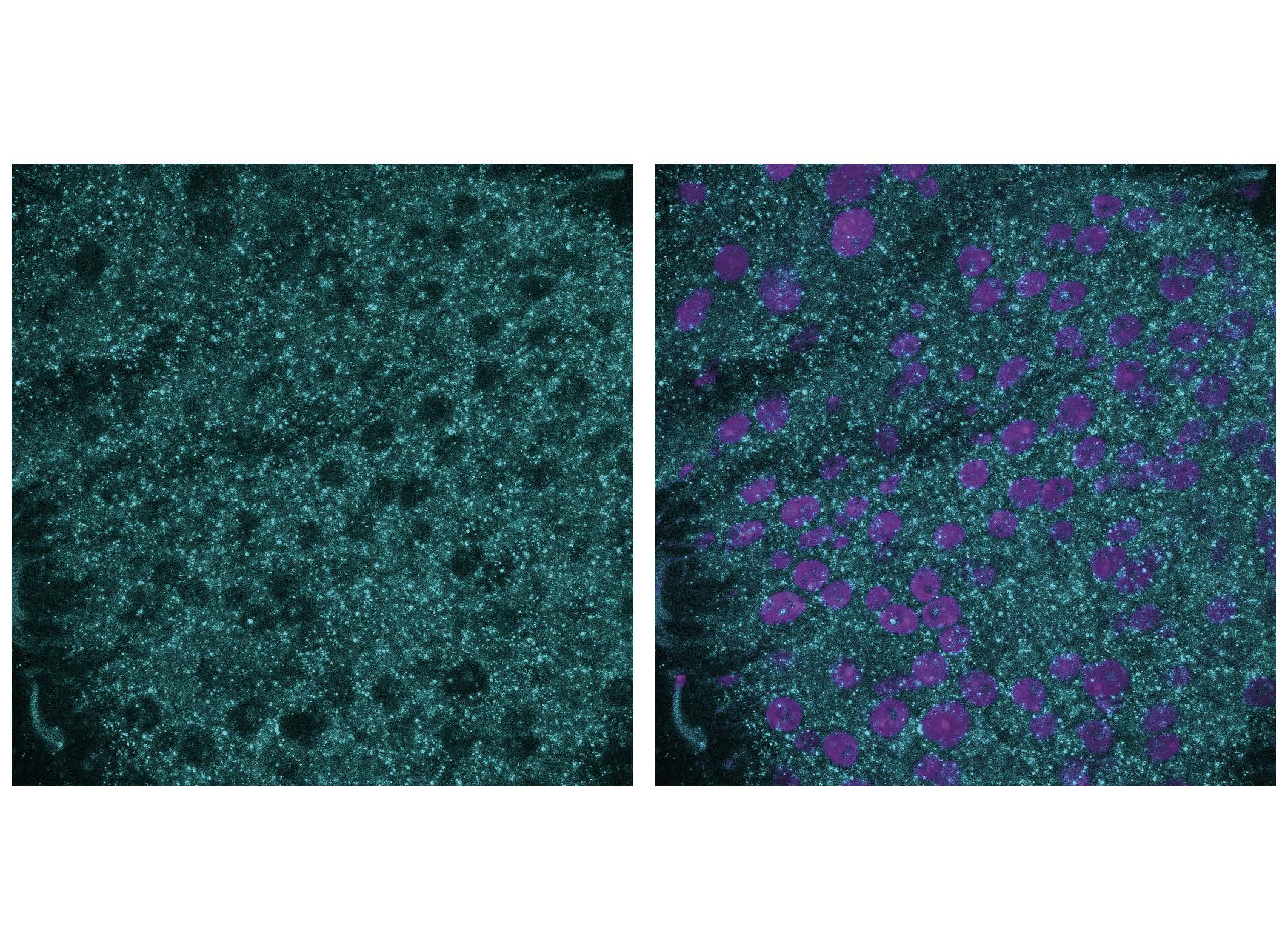
To date, only a single enzyme has been identified and characterized in eukaryotes that ensures the post-translational repair of iso-L- and D-aspartate residues derived from L-aspartate. We combine interdisciplinary chiral-selective detection tools with reverse genetic screening to identify new chirality-regulating genes specialized in repairing non-L-amino acid residues, thereby preserving proteome homochirality.
Figure 5. Immunostaining of the chiral-deficient adult midgut with heterochiral neoepitope-specific antibodies developed by our team.
Recent publications
- Banreti, A, Bhattacharya, S, Wien, F, Matsuo, K, Réfrégiers, M, Meinert, C et al.. Biological effects of the loss of homochirality in a multicellular organism. Nat Commun. 2022;13 (1):7059. doi: 10.1038/s41467-022-34516-x. PubMed PMID:36400783 PubMed Central PMC9674851.
- Klionsky, DJ, Abdel-Aziz, AK, Abdelfatah, S, Abdellatif, M, Abdoli, A, Abel, S et al.. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition)1. Autophagy. 2021;17 (1):1-382. doi: 10.1080/15548627.2020.1797280. PubMed PMID:33634751 PubMed Central PMC7996087.
- Duffraisse, M, Paul, R, Carnesecchi, J, Hudry, B, Banreti, A, Reboulet, J et al.. Role of a versatile peptide motif controlling Hox nuclear export and autophagy in the Drosophila fat body. J Cell Sci. 2020;133 (18):. doi: 10.1242/jcs.241943. PubMed PMID:32878938 .
- Banreti, AR, Meier, P. The NMDA receptor regulates competition of epithelial cells in the Drosophila wing. Nat Commun. 2020;11 (1):2228. doi: 10.1038/s41467-020-16070-6. PubMed PMID:32376880 PubMed Central PMC7203100.
- Meier, P, Banreti, A. Tissue Repair: How to Inflame Your Neighbours. Curr Biol. 2016;26 (5):R192-4. doi: 10.1016/j.cub.2016.01.033. PubMed PMID:26954436 .
- Banreti, A, Hudry, B, Sass, M, Saurin, AJ, Graba, Y. Hox proteins mediate developmental and environmental control of autophagy. Dev Cell. 2014;28 (1):56-69. doi: 10.1016/j.devcel.2013.11.024. PubMed PMID:24389064 .
- Bánréti, A, Sass, M, Graba, Y. The emerging role of acetylation in the regulation of autophagy. Autophagy. 2013;9 (6):819-29. doi: 10.4161/auto.23908. PubMed PMID:23466676 PubMed Central PMC3672293.
- Bánréti, Á, Lukácsovich, T, Csikós, G, Erdélyi, M, Sass, M. PP2A regulates autophagy in two alternative ways in Drosophila. Autophagy. 2012;8 (4):623-36. doi: 10.4161/auto.19081. PubMed PMID:22330894 .
2024 - ANR JCJC Laureate
2023 - Lendulet Momentum Laureate
2015 - EMBO-Marie Curie Postdoctoral Fellow
iBV - Institut de Biologie Valrose
"Centre de Biochimie"
Université Nice Sophia Antipolis
Faculté des Sciences
Parc Valrose
06108 Nice cedex 2

